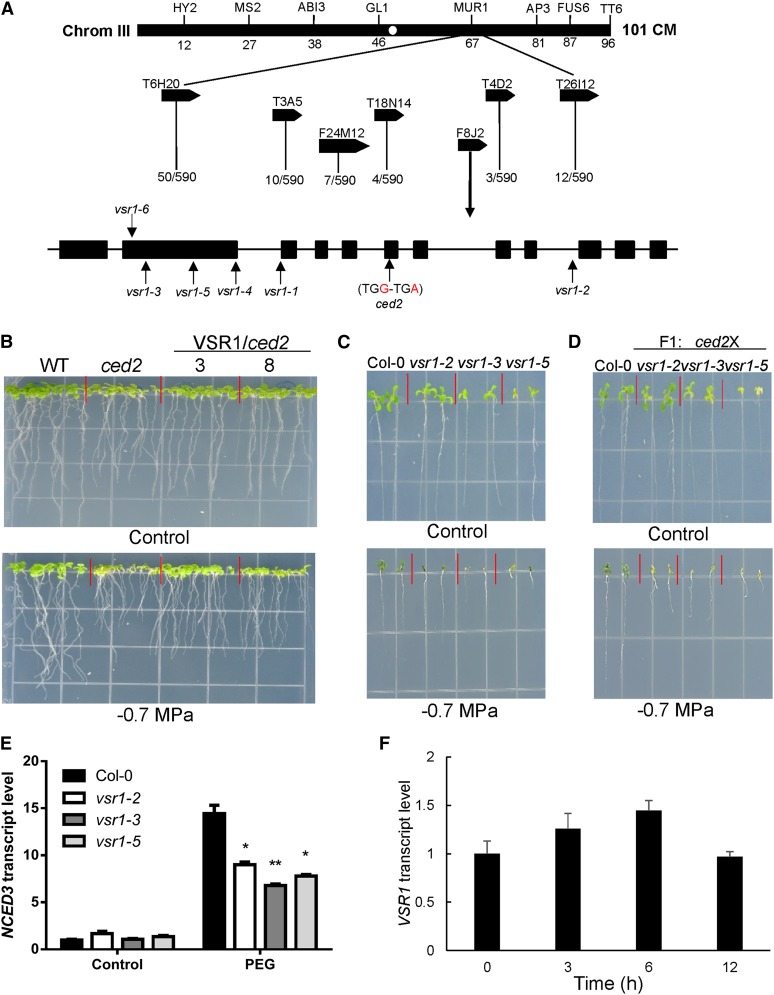
Figure 2.
Map-based cloning and complementation of the ced2 mutation and the expression of VSR1 under osmotic stress treatments. A, By analyzing 590 recombinant chromosomes, the CED2 locus was mapped to the lower arm of chromosome 3 between the bacterial artificial chromosome clones F8J2 and T4D2. DNA sequencing revealed that the ced2 mutant had a point mutation in the sixth exon of the At3g52850 gene. Positions of mutations in each allele are indicated by vertical arrows. B, Complementation of ced2 by the wild-type CED2/VSR1 gene. The wild type (WT), ced2, and two homozygous ced2 transgenic lines (nos. 3 and 8) expressing the wild-type CED2 gene are shown. Plants were grown on MS (Control, top) or –0.7 MPa PEG-infused agar plates (bottom). The photographs were taken 10 d after seed imbibition. C, vsr1 mutant alleles were sensitive to osmotic stress. Seeds were germinated, and seedlings were grown on control (top) or –0.7 MPa PEG-infused MS agar plate (bottom) for 2 weeks before taking the pictures. D, The ced2 mutant is a new allele of VSR1. Wild-type and F1 plants (ced2/vsr1-2, ced2/vsr1-3, and ced2/vsr1-5) were grown on MS (Control, top) or –0.7 MPa PEG-infused agar plate (bottom) for 2 weeks before taking the pictures. E, Transcript levels of NCED3 in the wild type and three vsr1 mutant alleles under the control or 40% (w/v) PEG treatment for 6 h. F, Regulation of CED2/VSR1 expression by osmotic stress. RNA was extracted from 2-week-old wild-type ecotype Columbia (Col-0) plants treated with 40% (w/v) PEG solution for the indicated time and was used for qRT-PCR analysis. In E and F, real-time reverse transcription-PCR quantifications were normalized to the expression of Polyubiquitin3 (UBQ3). Error bars represent se from three biological replicates. *P < 0.05 and **P < 0.01.