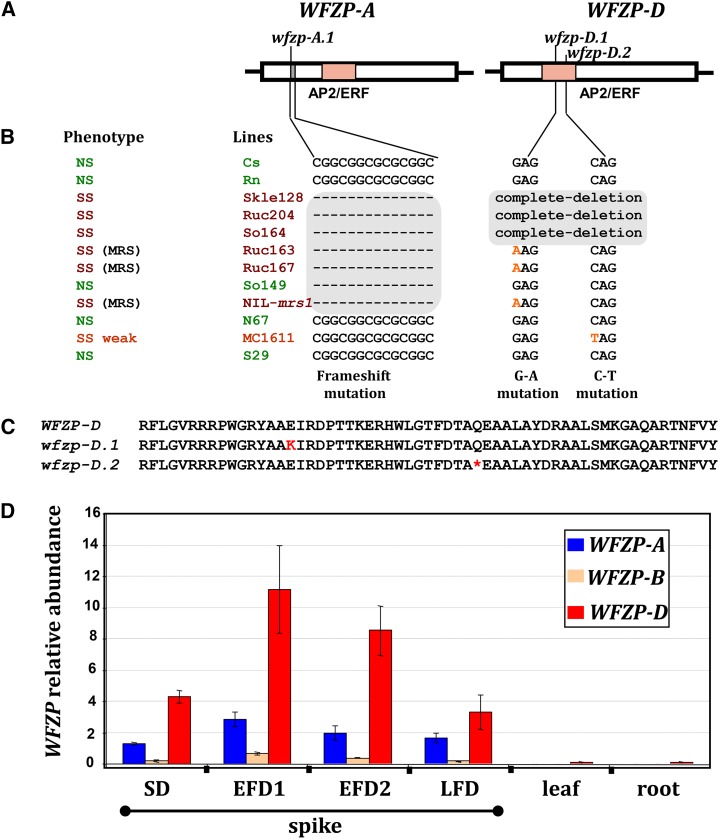
Figure 3.
Structural and functional characterization of WFZP genes. A, Schematic representation of the WFZP-A and WFZP-D gene structures and the mutations identified. The light red box indicates the AP2/ERF domain, and gray boxes indicate deletions. B, WFZP haplotypes of SS (red) and standard spiked (normal spiked [NS], green) lines. C, Alignment of the amino acid sequences of the AP2/ERF domains of wild type, WFZP-D, and wfzp-D.1, wfzp-D.2 mutants. Black asterisks indicate amino acids that confer specificity to the GCC-box binding site. D, Relative abundance of WFZP homoeolog mRNAs as determined by quantitative reverse transcription (qRT)-PCR with samples from spikes of bread wheat ‘Chinese Spring’ at various developmental stages of the spike (see below) and from roots and leaves. The values shown correspond to the mean values for three biological and three technical replicates, normalized to the value for the Ta.304.3 gene used as a reference. Error bars represent ses between replicates. Spike developmental stages were assigned as follows: spikelet differentiation stage (SD; when spikelets are differentiated to form two opposite rows at the rachis and glume primordia are initiated), early floret differentiation stage 1 (EFD1; spikelet meristems initiate floral meristems and lemma primordia are apparent), early floret differentiation stage 2 (EFD2; floral organ primordia are apparent in basal florets), and late floret differentiation stage (LFD; floral organs are differentiated in each floret).