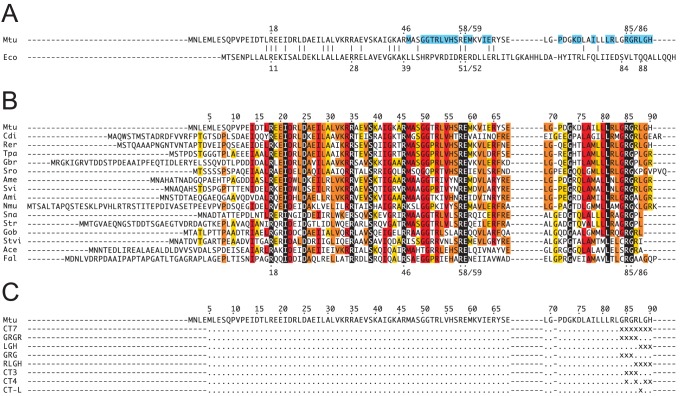
Figure 3. Sequence alignments of relevant AroQ chorismate mutases.
(A) Structural alignment based on an overlay of X-ray structures of EcCM (PDB: 1ECM) and MtCM (PDB: 2W1A) [10]. Catalytic residues are indicated with dots and numbers above or below the primary sequence. Residues that could assume the roles of EcCM’s Ser84 and Gln88 are missing in MtCM. MtCM residues within a 6-Å shell of MtDS are highlighted in cyan. (B) Multiple sequence alignment of representative AroQδ CMs from the order of Actinomycetales. The conservation of individual residues is color-coded by text highlighting in black, as 100%; red, ≥75%; orange, ≥50%; yellow, ≥33%; white, <33% identity; numbering according to the MtCM (Mtu) sequence. Abbreviations: Mtu, M. tuberculosis; Eco, E. coli; Cdi, Corynebacterium diphtheriae; Rer, Rhodococcus erythropolis; Tpa, Tsukamurella paurometabola; Gbr, Gordonia bronchialis; Sro, Segniliparus rotundus; Ame, Amycolatopsis mediterranei; Svi, Saccharomonospora viridis; Ami, Actinosynnema mirum; Nmu, Nakamurella multipartita; Sna, Stackebrandtia nassauensis; Str, Salinispora tropica; Gob, Geodermatophilus obscurus; Stvi, Streptomyces viridochromogenes; Ace, Acidothermus cellulolyticus; Fal, Frankia alni. (C) Up to seven C-terminal residues of MtCM were randomized in libraries CT7, GRGR, LGH, GRG, RLGH, CT3, CT4, and CT-L. Randomized positions are indicated as “x”, wild-type residues as dots.