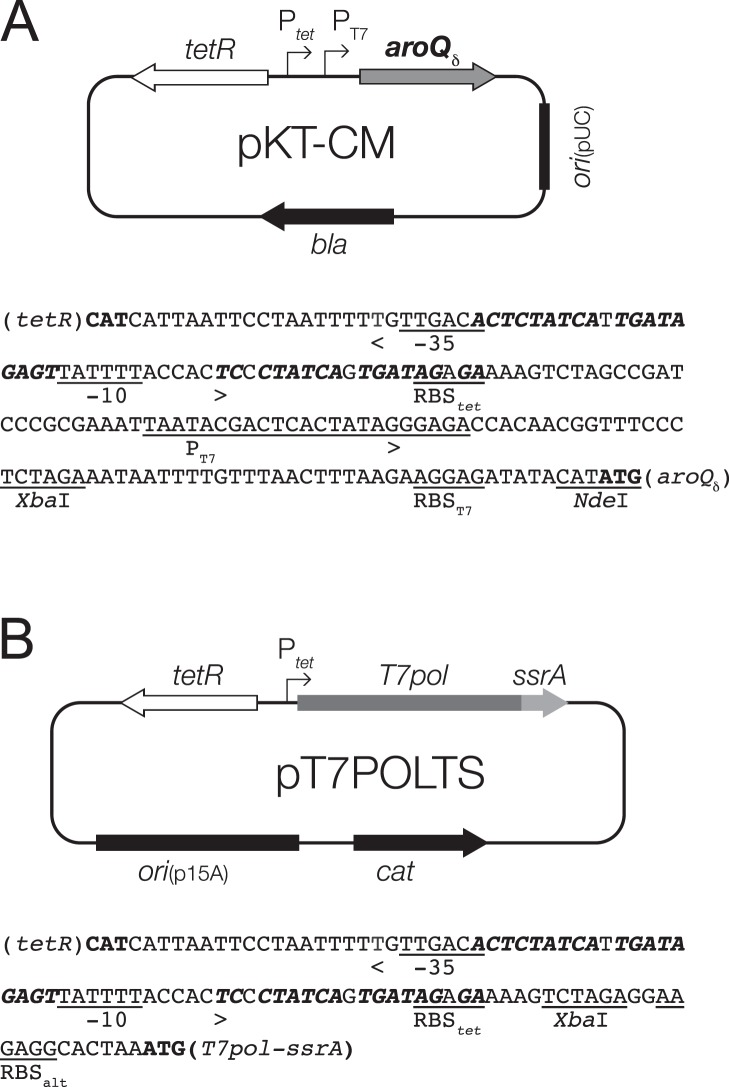
Figure 7. Overview of the plasmid-based T7 RNA polymerase gene expression system.
(A) Map and relevant promoter sequence of library plasmid pKT-CM. This plasmid was used for both in vivo selection (relying on Ptet, see also Fig. 4B) and in vitro overproduction of the MtCM variants (using PT7). The sequence of the PtetPT7 tandem promoter is given with the binding sites for the Tet-responsive TetR repressor highlighted in bold italics and the start codons of the reading frames in bold roman type [49]; underlined are relevant restriction sites, the ribosomal binding site (RBStet), −35 and −10 regions of Ptet [49] and the RBST7 and promoter PT7 from phage T7 [23], [50]; start and direction of transcription is marked by an arrowhead [22]. (B) Map and relevant promoter sequence of the T7 RNA polymerase plasmid pT7POLTS. The plasmid carries the p15A origin of replication derived from pACYC184 [51], cat encoding chloramphenicol acetyltransferase, and a Ptet controlled T7 RNA polymerase gene (T7pol) translationally fused at its 3′ end to the sequence for the SsrA degradation tag. In the absence of Tet, TetR binding to its operator sites (highlighted as in panel A) blocks gene expression from Ptet and any T7 RNA polymerase produced due to low-level leaky transcription is effectively eliminated by SsrA-mediated Clp proteolysis, thereby suppressing basal polymerase activity. Provision of Tet releases TetR from the operator, resulting in intracellular polymerase levels higher than can be degraded efficiently by the Clp proteases [41]. For efficient translation, the alternative RBSalt can be used. The accumulating polymerase then directs massive transcription from PT7 controlled genes, such as aroQδ on pKT-CM. The entire nucleotide sequence of pT7POLTS is provided as S2 Fig.