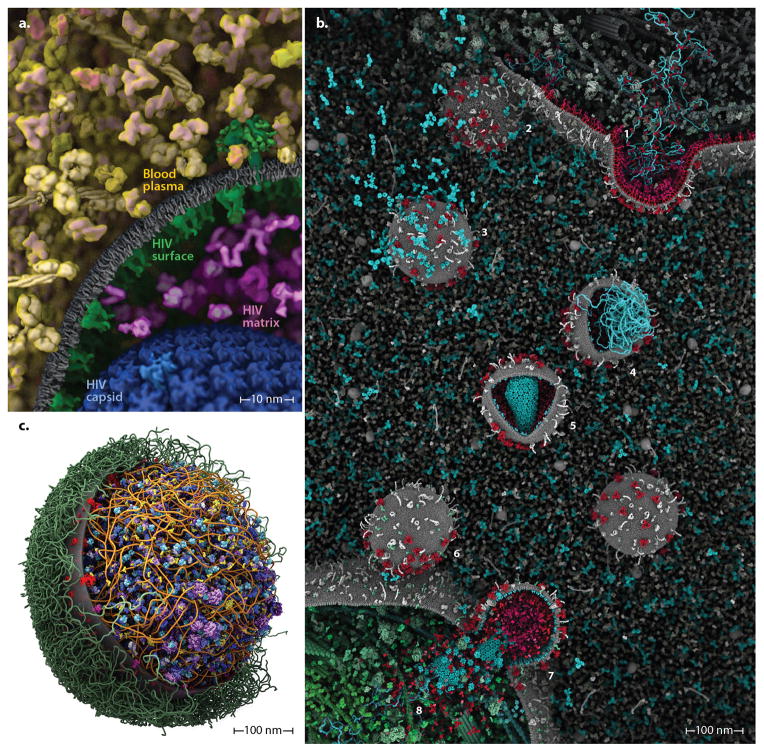
Figure 6.
cellPACK can integrate recipes into unified hybrid models that may be iteratively refined by content experts and systems-biology database improvements for each organelle system. (a) An HIV-in-BloodPlasma model shows seven different recipes (each a different color coding) unified into a single model. (b) As part of the autoPACK Visualization Challenge, J. Klusak generated this complete model of the HIV life cycle (including an animated version of most components) using the cellPACK software in conjunction with molecular modeling packages mMaya (http://www.molecularmovies.com/toolkit) and Chimera to generate additional molecular ingredients (see http://www.cgsociety.org/index.php/CGSFeatures/CGSFeatureSpecial/autopack_challenge_winners). (c) Our first model of a whole cell, Mycoplasma mycoides integrates recipes with surface, fibrous, and volumetric ingredients into a single comprehensive model. Interactive versions, updates and variations of these models can be viewed online at http://www.autopack.org/gallery/autofill-viewers.