Figure 2.
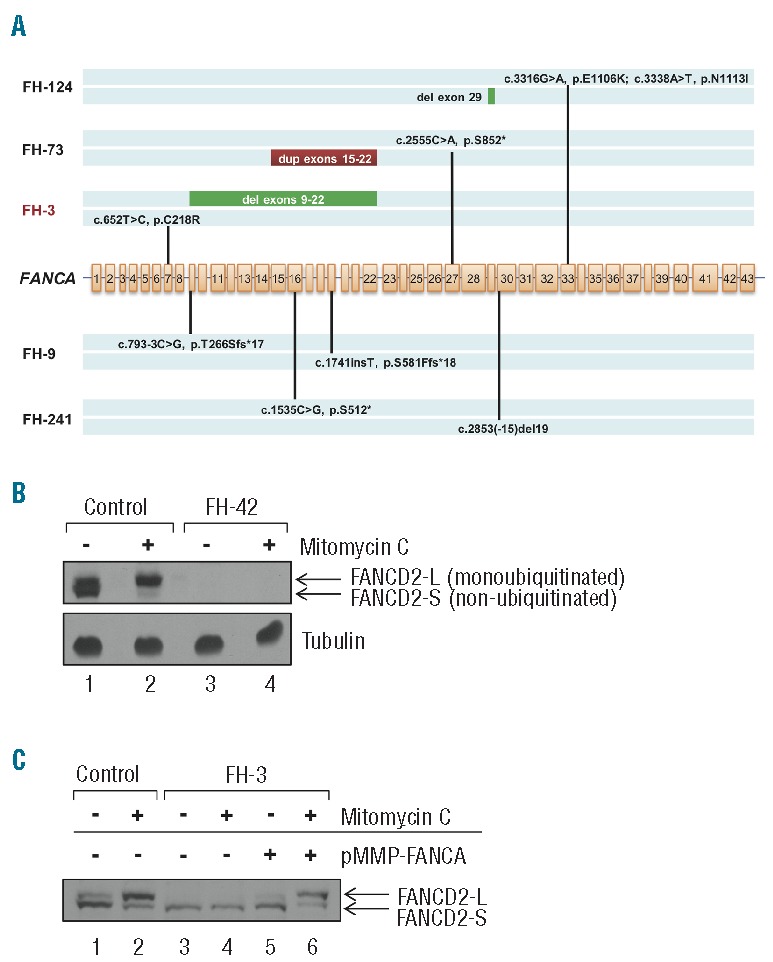
Targeted gene capture correction of Fanconi anemia subtype assignment. (A) Biallelic FANCA mutations identified by MarrowSeq in 5 patients. FA patient FH-3 (highlighted in red) was non-ACG subtype by clinical complementation testing. (B) Protein extracts of bone marrow fibroblasts isolated from healthy controls or Fanconi anemia patient FH-42 (FANCD2, p.[Leu683Pro];[Glu906Ilefs*4]) were immunoblotted for FANCD2 with or without 24-h mitomycin C treatment. Fibroblasts from FH-42 exhibit low FANCD2 protein expression (lanes 3 and 4) in comparison to cells from controls (lanes 1 and 2). α-tubulin was used to ascertain equivalent protein loading. (C) Functional validation of Fanconi anemia subtype A in FA patient FH-3 (FANCA p.[Cys218Arg];[Val265Leufs*8]). Protein extracts of bone marrow fibroblasts isolated from healthy control or FH-3 were immunoblotted for FANCD2 with or without 24-h mitomycin C treatment. Fibroblasts from healthy control show both non-ubiquitinated (FANCD2-S) and monoubiquitinated (FANCD2-L) FANCD2 forms (lane 1), with an increased ratio of monoubiquitinated FANCD2 relative to non-ubiquitinated FANCD2 upon mitomycin C treatment (lane 2). Fibroblasts from FH-3 show only the non-ubiquitinated FANCD2-S form with and without mitomycin C (lanes 3 and 4). FANCD2 monoubiquitination is restored upon infection with a pMMP retroviral vector encoding the wild-type FANCA cDNA (lanes 5 and 6).
