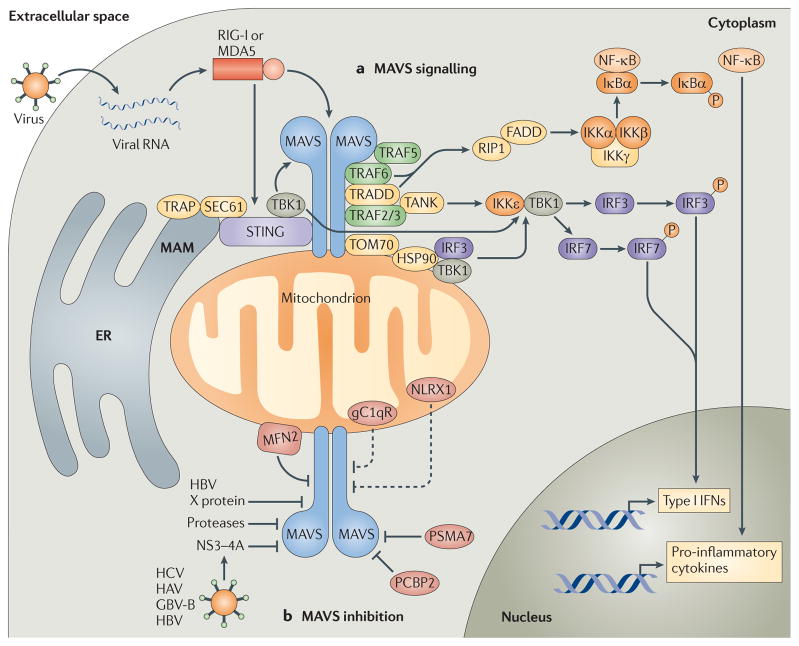
Figure 1. Mitochondrial antiviral signalling pathways.
a | Cytosolic viral RNA is recognized by the RIG-I-like receptors (RLRs) retinoic acid-inducible gene I (RIG-I) and melanoma differentiation-associated gene 5 (MDA5), which activate mitochondrial antiviral signalling protein (MAVS) through caspase-recruitment domain (CARD)–CARD interactions. MAVS then recruits various signalling molecules to transduce downstream signalling, such as TNF receptor-associated factor 6 (TRAF6) and TRAF5. TRAF6, along with TNFR1-associated death domain protein (TRADD), activates canonical nuclear factor-κB (NF-κB) signalling via receptor-interacting protein 1 (RIP1) and FAS-associated death domain protein (FADD). Canonical NF-κB signalling occurs as the IκB kinase (IKK) complex — consisting of IKKα, IKKβ and IKKγ — phosphorylates NF-κB inhibitor-α (IκBα), resulting in the proteasomal degradation of IκBα and thus liberating NF-κB to translocate into the nucleus and initiate pro-inflammatory cytokine gene expression. MAVS also interacts with various molecules that activate interferon regulatory factor (IRF) signalling (such as stimulator of interferon genes (STING)). These molecules, together with the translocon-associated protein (TRAP) complex and the SEC61 translocon, mediate the activation of TANK-binding kinase 1 (TBK1), which phosphorylates IRF3 and IRF7. In addition, MAVS interacts with translocase of the outer membrane 70 (TOM70), which also interacts with heat shock protein 90 (HSP90) and thereby localizes TBK1 and IRF3 in proximity to the MAVS signalosome. Finally, MAVS binds TRAF2 and TRAF3 and, through TRADD and TRAF family member-associated NF-κB activator (TANK) interactions, promotes IKKε- and/or TBK1-mediated phosphorylation of IRF3. This promotes IRF3 nuclear translocation, leading to the expression of type I interferon (IFN) genes. b | MAVS signalling can also be inhibited by various molecules. For example, hepatitis C virus (HCV) encodes a serine protease, termed NS3–4A, that inhibits MAVS by cleaving it from the outer mitochondrial membrane and preventing the formation of MAVS–IKKε signalling complexes. Hepatitis A virus (HAV) and GB virus B (GBV-B) also encode proteases that disrupt the mitochondrial targeting of MAVS, and hepatitis B virus (HBV) X protein was shown to promote polyubiquitin conjugation to MAVS, leading to its degradation. Endogenous molecules such as poly(rC)-binding protein 2 (PCBP2) and the 20S proteasomal subunit PSMA7 can negatively regulate MAVS signalling during viral infection by promoting its degradation. Other molecules, such as mitofusin 2 (MFN2), receptor for globular head domain of complement component 1q (gC1qR) and NLR family member X1 (NLRX1) are also thought to inhibit MAVS signalling by direct interaction, although gC1qR and NLRX1 are thought to localize predominately to the mitochondrial matrix. Therefore, the mechanisms by which these molecules inhibit MAVS signalling remain under investigation (dashed lines). ER, endoplasmic reticulum; MAM, mitochondria-associated membrane.