Abstract
The 22q11.2 deletion syndrome (22q11DS) is thought to be a contiguous gene syndrome caused by haploinsufficiency for a variable number of genes with overlapping function during the development of the craniofacial, pharyngeal and cardiac structures. The complexity of genetic and developmental anomalies resulting in 22q11DS has made attributing causation to specific genes difficult. The CRKL gene resides within the common 3-Mb region, most frequently affected in 22q11DS, and has been shown to play an essential role in the development of tissues affected in 22q11DS. Here, we report the characterisation of a mouse strain we named ‘snoopy’, harbouring a novel Crkl splice-site mutation that results in a loss of Crkl expression. The snoopy strain exhibits a variable phenotype that includes micrognathia, pharyngeal occlusion, aglossia and holoprosencephaly, and altered retinoic acid and endothelin signalling. Together, these features are reminiscent of malformations occurring in auriculocondylar syndrome and agnathia-otocephaly complex, 2 conditions not previously associated with the CRKL function. Comparison of the features of a cohort of patients harbouring small 22q11.2 deletions centred over the CRKL gene, but sparing TBX1, highlights the role of CRKL in contributing to the craniofacial features of 22q11DS. These analyses demonstrate the central role of Crkl in regulating signalling events in the developing oropharyngeal complex and its potential to contribute to dysmorphology.
Key Words: Craniofacial development, Crkl, Deletion 22q11.2, Malformation, Pharyngeal patterning
Deletions of the human chromosome 22q11.2 are common and result in a complex syndrome characterised by distinctive craniofacial malformations such as a long tubular nose, micrognathia and a cleft palate as well as congenital heart disease and other anomalies (also known as velocardiofacial syndrome, see OMIM 192430, 188400) [Tan et al., 2010]. The pathogenesis of 22q11.2 deletion syndrome (22q11DS) is complex and has been the subject of many human genetics and animal studies. TBX1 and CRKL localize to the major 3-Mb region most commonly deleted in 22q11DS. Point mutations of TBX1 in 22q11DS-like individuals and Tbx1 mouse models strongly implicate this gene as the major phenotypic determinant of 22q11DS. However, many patients with a phenotype ostensibly indistinguishable from 22q11DS have atypical deletions not involving TBX1 [Kurahashi et al., 1997; Garcia-Miñaur et al., 2002; Rauch et al., 2005; Fernández et al., 2009; Ogilvie et al., 2009] or 2 normal copies of TBX1 [Gong et al., 2001; Conti et al., 2003], indicating that a loss of TBX1 function is not the sole mechanism responsible for the developmental anomalies associated with 22q11DS [Gong et al., 2001; Conti et al., 2003]. The analysis of Tbx1;Crkl compound heterozygous mice revealed an interaction between these two 22q11.2 genes during pharyngeal development, suggesting that 22q11DS is a contiguous gene disorder and implicated altered retinoic acid signalling as an underlying cause of abnormal pharyngeal development [Guris et al., 2006]. Atypical 22q11.2 microdeletions that encompass CRKL, but spare TBX1, have been demonstrated in a number of individuals [Breckpot et al., 2012; Verhagen et al., 2012], suggesting that a loss of CRKL alone may be responsible for 22q11DS phenotypes in some cases.
Crkl is an SH2-SH3 domain adaptor protein with roles in propagating signalling from growth factor systems including Fgf [Moon et al., 2006], oestrogen [Padmanabhan et al., 2011] and TGFβ [Wurdak et al., 2005; Lv et al., 2013] which mediate a range of cellular activities including proliferation, migration and adhesion. Homozygous deletion of Crkl in mice results in late-gestation lethality and a range of developmental defects that recapitulates many of the features of 22q11DS. These include a range of cardiac malformations such as double-outlet right ventricle and ventricular septal defects, defects in thymus and parathyroid development and craniofacial malformation [Guris et al., 2001, 2006]. The features of 22q11DS and the Crkl null mouse are suggestive of a neurocristopathy, implying that Crkl-mediated signalling is important for neural crest cell development.
We have identified an ethylnitrosourea (ENU) mutant mouse strain, which we named ‘snoopy’ [Caruana et al., 2013], harbouring a point mutation in the Crkl gene. The snoopy strain exhibits severe craniofacial malformation, including maxillary and mandibular hypoplasia, microstomia, aglossia, pharyngeal occlusion, and holoprosencephaly with markedly variable expressivity and reduced penetrance. The snoopy mutation induces aberrant splicing of Crkl resulting in the production of a mutant transcript and a loss of Crkl protein. Expression analysis reveals that both retinoic acid and endothelin signalling are increased in snoopy embryos. We also present a cohort of patients with heterozygous chromosome 22q11.2 deletions involving CRKL, but not TBX1. The human phenotype is variable, but we have observed craniofacial anomalies such as micrognathia, a cleft lip and palate and a deviated nasal septum. The presumed mechanism underlying the human deletion phenotype is that of haploinsufficiency. Together, these phenotypic and molecular data suggest that snoopy models major phenotypic features seen in a range of craniofacial malformation syndromes, strengthens its relationship to the craniofacial anomalies observed in patients with atypical 22q11DS and highlights the central role of Crkl in regulating a network of genes essential for normal craniofacial development.
Materials and Methods
Mice
The identification of snoopy embryos from a large-scale ENU mutagenesis programme has been reported previously [Caruana et al., 2013]. All mouse procedures were approved by the Royal Children's Hospital Animal Ethics Committee, RCH AEEC No. A647 and A728. Embryos and mice were culled/harvested according to MCRI SOP No. 21.
Exome Enrichment, Sequencing, Variant Detection and Validation
The snoopy strain was maintained on an inbred C75BL6/c background. Generation of snoopy progeny, DNA extraction from affected snoopy embryos and exome enrichment was performed as described previously [Andrews et al., 2012; Caruana et al., 2013].
PCR, Sequencing and Genotyping
PCR, sequencing and high-resolution melt genotyping have all been described previously [Miller et al., 2013a]. Gene-specific primers were designed for DNA amplification of the exon/intron boundary of exon 1 in the murine Crkl gene (NC_000082.6). High-resolution melt analysis of Crkl exon 1 and intron 1 donor site was performed using primers Crkl.genF: 5′-CCTGGACACTACCACCTTAATC-3′ and Crkl.genR: 5′-ACCGGAGAAGGGCAGGAG-3′.
In situ Probe Synthesis and Whole-Mount in situ Hybridisation
The production of in situ hybridisation probes and whole-mount in situ hybridisation techniques have been detailed previously [Miller et al., 2013a, 2013b]. The images are representative of a minimum of 3 independent embryos per genotype.
Immunoblotting and Quantitative Real-Time RT-PCR
Whole E10.5 embryos were collected individually and sonicated in either Laemmli SDS Sample Buffer plus 0.1 M DTT or Trizol reagent (Life Technologies). For immunoblotting, 100 μg of protein for each genotype was run through a 12% Tris-Glycine gel, probed with a rabbit anti-CRKL antibody raised against an N-terminal epitope (1:1,000; LifeSpan Biosciences) as described [Miller et al., 2013a]. qPCR reactions were performed in triplicate for Crkl, Plcb4, Gnai3, Dlx5, Dlx6, and Hand2 genes on a Lightcycler 480 System using GoTaq qPCR Master Mix (for primers, see table 1). Expression levels were normalised to the housekeeping gene eukaryotic translation elongation factor 2 (Eef2) using primers reported previously [Williams et al., 2013].
Table 1.
Primers used for qPCR reactions
| Gene | Forward primer (5′-3′) | Reverse primer (5′-3′) |
|---|---|---|
| Crkl | TCCAGAAGAGAGTACCTTGTGC | GCATGTGACAGAGTGCTTCC |
| Plcb4 | AACGAACATCAGCGAGATCC | AGGTGTTGTGGGAGGAACTG |
| Gnai3 | GAGAAAGCGGCCAAAGAAG | TAACTGTCGGGCATCATCTG |
| Dlx5 | GGCTACTGCTCTCCTACCTCTG | TTCACCATCCTCACCTCTGGC |
| Dlx6 | GACTCAATACCTGGCCCTTCC | GGTCACTCTCGTGTGGGTTA |
| Hand2 | CTGGCCACCAGCTACATCG | TTCTTGTCGTTGCTGCTCACT |
Cell Culture and Cycloheximide Treatment
Individual whole E10.5 embryos were disaggregated in 0.025% Trypsin, split into 2 wells of a 6-well dish and then cultured in DMEM containing 10% (v/v) FCS. The plates were incubated overnight at 37°C with 5% CO2. Then, 100 μg/ml cycloheximide was added to the medium of one well, incubated for 4 h at 37°C with 5% CO2, and RNA was extracted (Trizol) for cDNA synthesis as described previously [Miller et al., 2013a]. Amplification of cDNA was performed using the following primers: Crkl-F 5′-CTGACCAAGCCCTCTGATTG-3′ and Crkl-R 5′-GCATGTGACAGAGTGCTTCC-3′.
Human Patient Samples
Individuals with chromosome 22q11.2 deletions involving CRKL, but not TBX1, were identified through the Victorian Clinical Genetics Services. DNA was extracted from peripheral blood samples, and SNP microarray was performed using the Illumina HumanCytoSNP-12 version 2.1 as described previously [Bruno et al., 2011]. Consent was obtained from affected individuals or their parents for the use of photographs and radiographic images. All human analyses were approved by the Royal Children's Hospital Human Research Ethics Committee project 31190.
Results
A Crkl Mutation Is the Basis of the Snoopy Strain
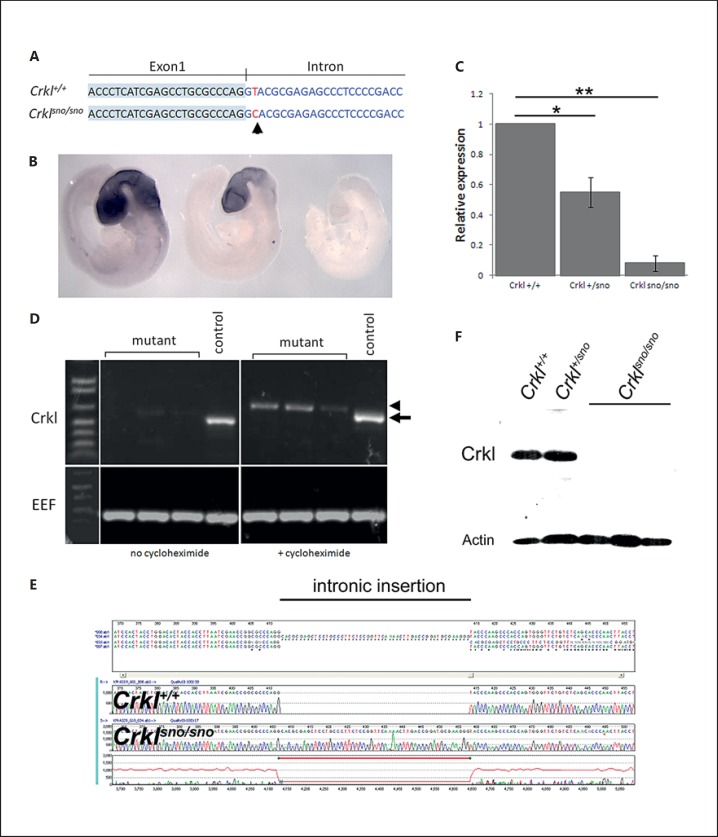
We performed an ENU mutagenesis screen in mice to identify genes with major roles in regulating embryonic development and which could, therefore, be candidates for causing human birth defects [Caruana et al., 2013]. Snoopy, one of the strains identified in this screen, exhibited a distinct but highly variable, recessively inherited, craniofacial phenotype involving micrognathia, holoprosencephaly and eye abnormalities. We utilized whole exome sequencing in 2 affected embryos to identify the causative mutation. We found 22 single nucleotide variants (SNVs) which were homozygous in both embryos sequenced and which could result in potentially deleterious consequences for the gene or its translated protein based on PolyPhen predictions. None of the 22 SNVs showed an absolute correlation with the phenotype. However, one of these SNVs, a T>C substitution in the Crkl gene (at position Chr16:17,452,790;GRCm38), was the only variant always homozygous in phenotypically mutant embryos, although a significant proportion of phenotypically normal embryos also harboured homozygous mutations. This T>C substitution resides 2 nucleotides downstream of exon 1 (fig. 1A), and the bioinformatic analysis of the mutant transcript predicted a loss of a putative splice donor and generation of additional exonic splice enhancers.
Fig. 1.
Identification of a Crkl mutation in snoopy mice. A Location of snoopy mutation (arrow) relative to Crkl coding sequence. B Whole-mount in situ hybridisation for the Crkl gene in wild-type, heterozygous and homozygous mutant embryos. C Relative quantitation of Crkl transcripts in snoopy embryos (* p = 0.0115, ** p = 0.0007). D Stabilisation of Crklsno transcript by cycloheximide. Cycloheximide treatment of embryonic fibroblasts results in stabilisation of the mutant RNA (arrowhead) which migrates more slowly than the wild-type transcript (arrow). EEF (eukaryotic translation elongation factor) was used as a loading control. E Sequencing the cycloheximide-stabilised mutant transcript revealed inclusion of 51 nucleotides of the Crkl intron. F Western blot analysis of protein extracted from whole E13.5 embryos demonstrated the loss of Crkl protein in mutants.
We, therefore, examined the expression of the Crkl transcript by whole-mount in situ hybridisation in genotypically defined embryos. This analysis demonstrated a loss of expression in both heterozygous and homozygous embryos relative to the wild type (fig. 1B), suggesting that the mutation destabilises the transcript, consistent with aberrant splicing. Quantitative RT-PCR demonstrated an approximate halving of transcript levels in Crkl+/sno embryos and a reduction to less than 10% of wild-type levels in Crklsno/sno embryos (fig. 1C). Conventional RT-PCR analysis of RNA extracted from embryonic fibroblasts resulted in a very weak amplification of Crklsno/sno mRNA relative to controls. Cycloheximide treatment of fibroblasts resulted in significantly greater amplification of a slightly larger transcript in mutant cells, suggesting stabilisation of an aberrant transcript (fig. 1D). Sequencing of this aberrant RT-PCR product revealed the insertion of 51 nucleotides of intronic sequence into the mature transcript, confirming the existence of aberrant splicing which results in mRNA degradation (fig. 1E). Western blot analysis verified the dramatic loss of Crkl protein in mutants, confirming the impact of Crkl mRNA degradation (fig. 1F). These data suggest that the snoopy strain arises from Crkl haploinsufficiency due to nonsense-mediated decay of the mutant transcript.
The Snoopy Phenotype Is Characterised by Distinct but Variable Craniofacial Anomalies
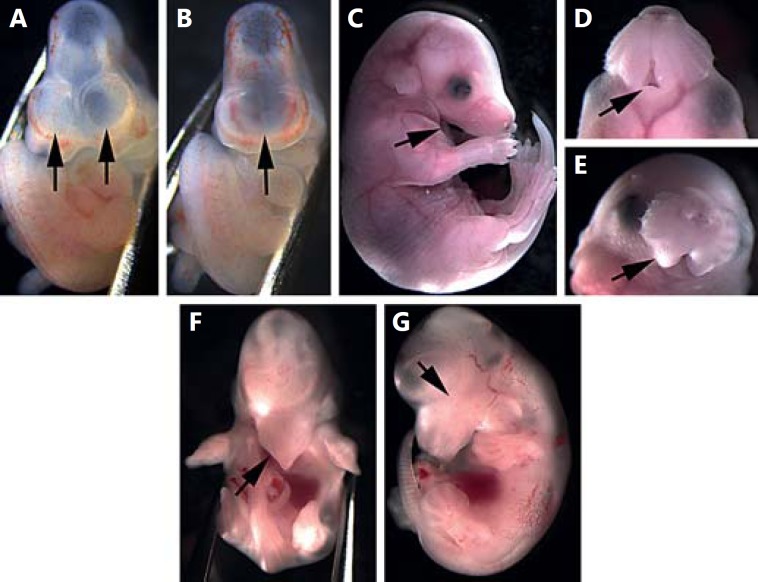
At E10.5, the paired forebrain vesicles are clearly apparent in controls, while mutant embryos frequently exhibit a single, small forebrain vesicle (fig. 2A, B). We also identified 2 embryos with overt cyclopia and a rostral proboscis (not shown). Typical mutants exhibit a variably affected mandible ranging from mild hypoplasia to agnathia (fig. 2C, D). Mandibular hypoplasia may also be accompanied by maxillary hypoplasia (fig. 2E, F). In more severely affected individuals, failure of maxillary and mandibular structures occurs in conjunction with microphthalmia or anophthalmia and holoprosencephaly (fig. 2F, G). While these craniofacial defects are frequently bilateral, a significant proportion of embryos exhibit asymmetric defects, particularly in the mandible.
Fig. 2.
Phenotype of snoopy embryos. At E10.5, the forebrain has divided into 2 distinct vesicles in control embryos (A), while snoopy embryos commonly exhibit a single, large forebrain vesicle (B). Snoopy embryos exhibit a range of orofacial malformations including mandibular hypoplasia and agnathia (C, D), maxillary hypoplasia (E, F) and occasional microphthalmia or anophthalmia (G).
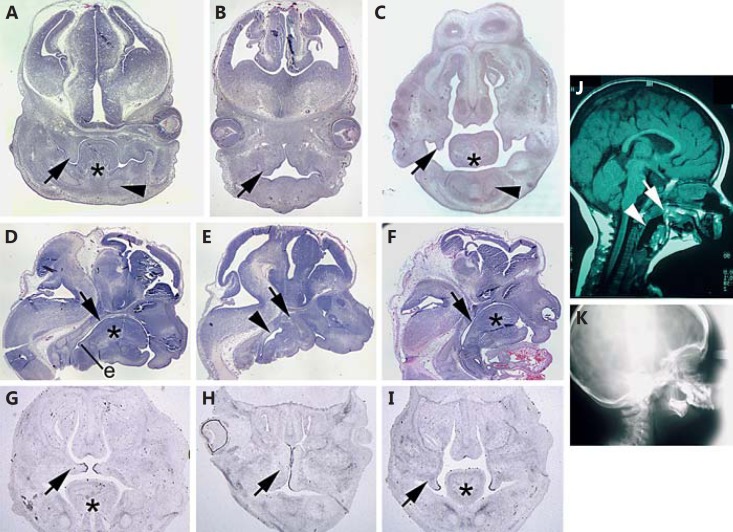
E13.5 snoopy embryos exhibited additional structural orofacial anomalies. Individuals with micrognathia exhibited aglossia or less commonly, severe hypoplasia of the tongue (fig. 3A, B). In embryos with aglossia, the palatal shelves were expanded and irregular in comparison to the narrow shelf-like morphology of controls (fig. 3B). In Crklsno/sno embryos with mild to no facial hypoplasia, the tongue appeared to be relatively normal, but palatal shelf elevation was delayed (fig. 3C). The complete failure of tongue development in severely affected mutants was confirmed by examining affected embryos in the sagittal plane. In addition, this demonstrated that the mandibular rudiment had undergone fusion with the roof of the oral cavity, resulting in complete occlusion of the pharynx (fig. 3D, E). Crklsno/sno embryos that developed a tongue did not exhibit occlusion of the pharynx (fig. 3F). Examination of the Tgfb3 expression in the palatal shelves at E14.5, immediately prior to normal fusion, revealed expression exclusively in the medial edge epithelium in wild-type controls (fig. 3G). Despite the grossly abnormal morphology of the palatal shelves in dysmorphic Crklsno/sno embryos, Tgfb3 was also expressed in the most medial region of the epithelium (fig. 3H). In Crklsno/sno embryos in which the tongue had formed, palatal shelf elevation is delayed, but Tgfb3 was expressed in a distribution indistinguishable from controls (fig. 3I). Severe micrognathia, mandibular hypoplasia, obstruction of the pharynx, and cleft palate are key phenotypic features of auriculocondylar syndrome (ACS, OMIM 602483, 614669). Comparison of the snoopy phenotype with clinical features of an individual with ACS reveals remarkable similarities including oropharyngeal occlusion and micrognathia (fig. 3J, K).
Fig. 3.
Oropharyngeal malformation in E13.5 snoopy embryos. In control E13.5 embryos (A), the vertical palatal shelves (arrow) are closely associated with the tongue, and Meckel's cartilage can be seen in cross section (arrowhead). In contrast, E13.5 snoopy mutants (B) fail to form a tongue, and the palatal shelves (arrow) are distended and abnormal in morphology. C In a Crklsno/sno embryo in which the tongue has formed, the palatal shelves take on a more typical morphology but are delayed in their elevation (arrow), and Meckel's cartilage is apparent (arrowhead). A sagittal section through the head of a control embryo (D) reveals the relationship of the tongue to the oral cavity. The oral cavity (arrow) is patent down to the epiglottis (e) and beyond into the larynx and opening of the oesophagus. A similar section in a snoopy embryo (E) demonstrates that there is no clear oral cavity and that the mandibular rudiment is continuous with the skull base (arrow). The pharynx distal to the occluded oral cavity (arrowhead) is typically distended relative to controls. A sagittal section through a mildly affected Crklsno/sno embryo (F) reveals mandibular hypoplasia and microglossia (asterisk) as well as a patent oral cavity (arrow). In situ hybridisation for Tgfb3 at E14.5 highlights the medial edge epithelium in the palatal shelves (arrow) of a wild-type embryo (G), in the dysmorphic palatal shelves of a severely affected Crklsno/sno embryo (H) or in the relatively normal but delayed palatal shelves of a mildly affected Crklsno/sno embryo (I). J Mid-sagittal computed tomography image of an individual with ACS illustrating a similar phenotype to snoopy including fusion of the mandible to the upper oropharynx (arrow) and a distended pharynx distal to the occlusion (arrowhead). K Lateral X-ray illustrating a diminutive mandible analogous to that observed in snoopy embryos. Asterisks indicate the tongue.
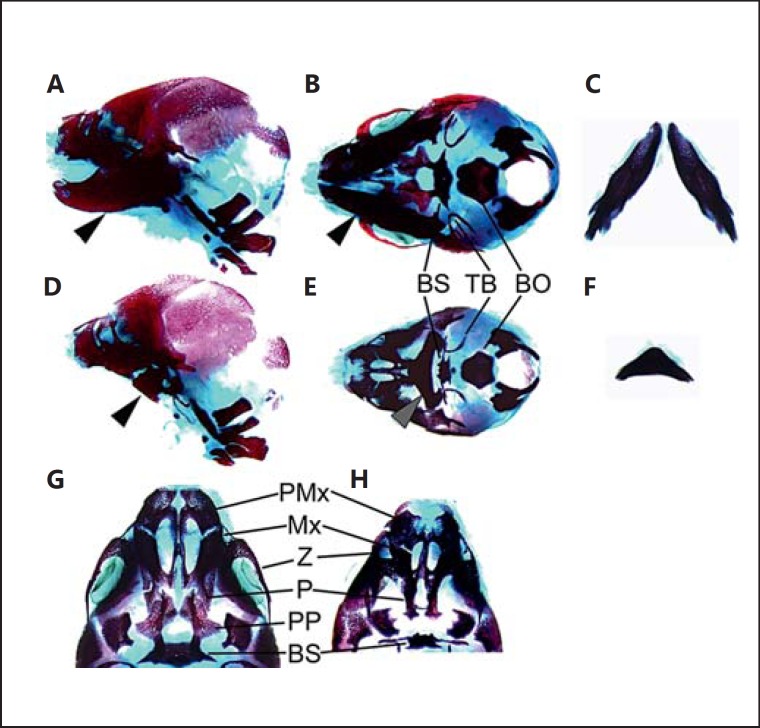
Analysis of skeletal preparations at E17.5 confirmed the extent of mandibular hypoplasia (fig. 4A–F). The mutant mandibular skeleton is severely dysmorphic, and the hemimandibles are prematurely fused distally (fig. 4C, F). The tympanic bullae are translocated medially towards a hypoplastic basisphenoid. The basisphenoid also harbours long, ectopic lateral processes that pass below, but are not fused to the tympanic bullae. The basioccipital appears relatively unaffected, but dissection of the mandible reveals a number of malformations anterior to the basioccipital (fig. 4G, H). The prepalatine bones are absent, and the palatine bones are reduced in length posteriorly. The zygomatic arch is reduced in size, and the maxilla/premaxilla are mildly hypoplastic.
Fig. 4.
Cranial skeletal morphology. Alizarin red and Alcian blue skeleton preparations of E17.5 wild-type control (A-C, G) and homozygous mutant Crklsno/sno (D-F, H) embryos. Lateral view of control (A) and mutant (D) illustrating the diminutive mandibular skeleton (arrowhead). Ventral view of control (B) and mutant (E) skull, and control (C) and mutant (F) isolated mandible. Ventral view of anterior skull base of control (G) and mutant (H) illustrating the impact of Crkl mutation on the palate and maxillary arch skeleton. The mutant skeleton preparation shown here (D-F, H) is from the same embryo shown in whole mount in figure 2 C-E. BS = Basisphenoid; TB = tympanic bulla; BO = basioccipital; PMx = pre-maxilla; Mx = maxilla; Z = zygomatic arch; P = palatine; PP = pre-palatine.
Retinoic Acid and Endothelin Signalling Pathways Are Perturbed in Snoopy Embryos
As an initial analysis of the molecular basis for the pharyngeal anomalies seen in snoopy embryos, we examined Cyp26a1 and Etv4 and 5 genes important for pharyngeal patterning and which respond to a loss of Crkl [Guris et al., 2006; Moon et al., 2006]. In situ hybridisation analysis revealed an upregulation of Cyp26a1 in Crklsno/sno E10.5 embryos (n = 3/4; fig. 5A, B), suggesting an increase in retinoic acid signalling. In contrast, no consistent difference was detected in the expression of Etv4 or 5 (fig. 5C, D; Etv4 not shown).
Fig. 5.
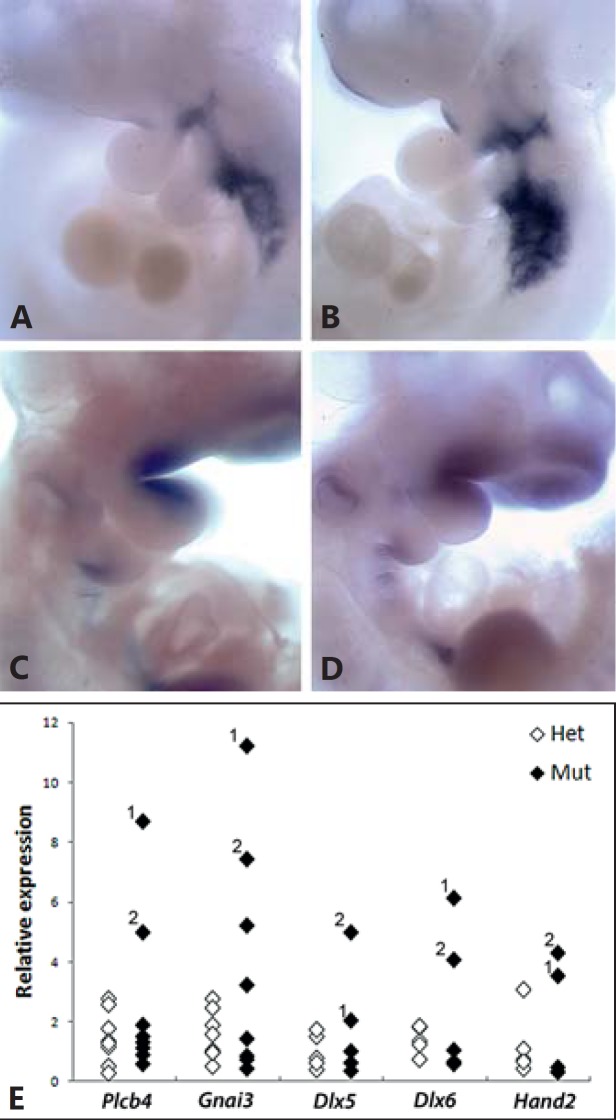
Altered expression of pharyngeal patterning genes in snoopy embryos. In situ hybridisation analysis of Cyp26a1 in Crkl+/+ (A) and Crklsno/sno (B) embryos. In situ hybridisation analysis of Etv5 in Crkl+/+ (C) and Crklsno/sno (D) embryos. E Real-time PCR analysis of gene expression in individual Crkl+/sno and Crklsno/sno embryos relative to Crkl+/+ embryos. Data points with same numbers are from the same embryos.
The recent identification of causative mutations in ACS and phenotypic overlap with snoopy embryos prompted us to examine the expression of Plcb4 and Gnai3. Expression levels were highly variable, but quantitative PCR analysis of RNA extracted from whole embryos revealed an increase in both Plcb4 and Gnai3 RNA levels in Crklsno/sno embryos relative to heterozygous and wild-type littermates (fig. 5E). Plcb4 and Gnai3 are hypothesized to mediate endothelin signalling during pharyngeal patterning through a network involving Hand2 and Dlx5 and 6. PCR analysis of whole embryo RNA demonstrated an elevated level of each of these 3 genes in the same embryos in which Plcb4 and Gnai3 expression changes were observed (fig. 5E), consistent with perturbed endothelin signalling following mutation of Crkl.
Variable Craniofacial Phenotype of Patients with CRKL – but Not TBX1 – Deletions
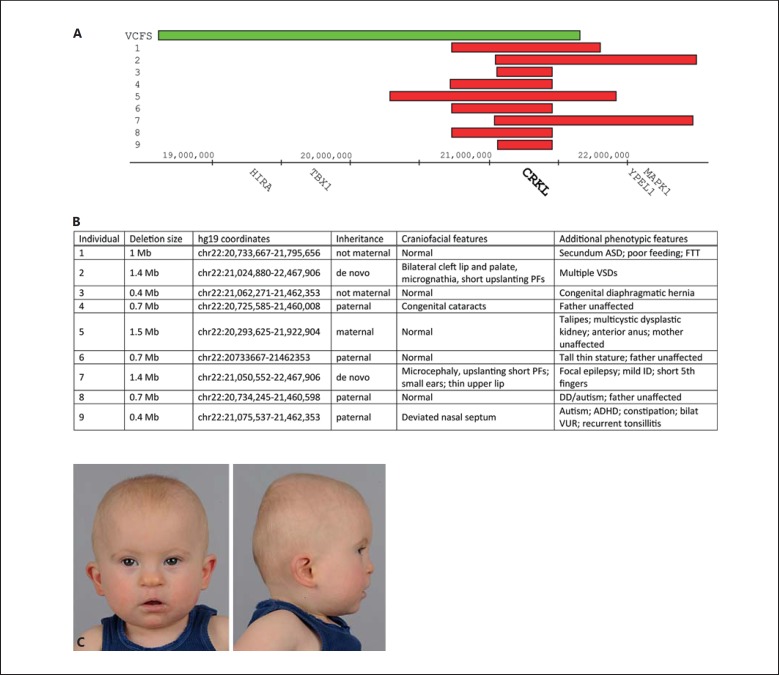
We sought to correlate our observations in the animal model with human disease and identified 9 probands with nested chromosome 22q11.2 deletions involving CRKL, but not TBX1 (fig. 6). All patients were ascertained because of developmental delay, congenital malformations, or both. Of all 9 deletions, 5 were inherited, 2 were of indeterminate inheritance (only one parent available for testing) and 2 were de novo. All 5 transmitting parents were phenotypically unaffected. In contrast to individuals with the typical large 3-Mb 22q11 deletion, individuals with the smaller nested CRKL deletion had a less consistently recognisable facial phenotype. Of our 9 probands, 2 (22%) exhibited significant craniofacial dysmorphism (fig. 6C). One had a bilateral cleft lip and palate and micrognathia; the other had a deviated nasal septum. None of our probands had the facial features usually observed in patients with the typical 22q11DS, such as a long tubular nose and hooded eyelids. The spectrum of congenital anomalies in our cohort is similar to that reported in 22q11DS, but there was striking phenotypic variability. Two individuals had cardiac septal defects, but the other malformations were observed in single individuals only. This is consistent with previous reports, which describe cardiac defects as the most common associated malformation [Verhagen et al., 2012]. We sequenced the nondeleted allele in patients 1-4 and 9, hypothesising that the individuals with craniofacial anomalies, particularly the individual with orofacial clefting and micrognathia, would harbour a CRKL variant that modified their phenotype. However, we found no significant coding variants in the nondeleted CRKL allele (data not shown).
Fig. 6.
Clinical and molecular details of individuals sharing heterozygous loss of CRKL. A Diagrammatic representation of nested chromosome 22q11.2 deletions in all 9 affected individuals (red bars) relative to the common 3-Mb deletion typically associated with velocardiofacial syndrome (VCFS; green bar). B Cytogenetic and clinical details of affected individuals and their parents. C Patient 2 illustrating distinctive facial features including low-set ears, upslanting palpebral fissures, a broad nasal bridge and tip, a flat mid-face and micrognathia. Note that the bilateral cleft lip and palate have been surgically repaired. ADHD = Attention deficit hyperactivity disorder; ASD = atrial septal defect; DD = developmental delay; FTT = failure to thrive; ID = intellectual disability; PFs = palpebral fissures; VSDs = ventricular septal defects; VUR = vesicoureteral reflux.
Discussion
In the present study, we show that an ENU-induced Crkl splice-site mutation results in a complex array of variable craniofacial malformation resembling a number of features observed in ACS. CRKL is also an important factor in the aetiology of the 22q11DS phenotype, and we present cases comparing the impact of a heterozygous loss of CRKL in humans. All together, these findings highlight the central role of Crkl in coordinating the development of the oropharyngeal complex and its potential to contribute to malformation when disrupted.
While there is a substantial overlap between the phenotype of snoopy and Crkl null mouse, snoopy embryos exhibit a number of phenotypes not reported in the Crkl null mouse including holoprosencephaly and dramatic oropharyngeal malformations. While the holoprosencephaly is difficult to reconcile with the reported Crkl null phenotypes, the oropharyngeal phenotypes could be more extreme manifestations of those reported previously. The reasons for these differences are not clear, but could be due to strain differences. Alternatively, since snoopy results from an in-frame splicing defect in the Crkl gene, these differences may reflect a residual level of mutant protein below the level of detection by Western blot. Further work will be required to determine if this is the case.
Considerable phenotypic variability is a common observation in many craniofacial malformation syndromes including ACS, and this may also manifest as incomplete penetrance [Gordon et al., 2013b]. This variation is often attributed to a heterogeneous genetic background involving the presence of particular modifying alleles. Our cohort of patients with chromosome 22q11.2 microdeletions involving CRKL, but not TBX1, also demonstrates marked clinical heterogeneity. However, only around half of all Crklsno/sno embryos have a phenotype detectable by gross examination despite their inbred background. Crkl is an adaptor protein with signal transduction roles in a number of major developmentally important pathways such as FGF and EGF signalling. It is possible that small stochastic variations at the level of growth factor signalling early in development could be amplified through subsequent developmental stages and result in substantial morphological differences between 2 genotypically identical embryos. Interestingly, the level of phenotypically abnormal snoopy embryos is similar to that reported for the Crkl null mouse [Guris et al., 2001], supporting the view that the variable penetrance may reflect inherent differences in the response to a loss of Crkl signalling between embryos.
Late-gestation snoopy embryos exhibit a cleft secondary palate, and at E13.5, immediately prior to palatal shelf elevation, embryos with aglossia also exhibit unusual palatal shelf morphology. The palatal shelves of mutant snoopy embryos are dramatically expanded and bulbous in appearance, filling most of the oral cavity. In mutants with aglossia, the concept of palatal shelf elevation appears inappropriate, since the shelves never extend vertically as such, nor do they have to navigate around the tongue. In Crklsno/sno embryos in which the tongue has formed, the palatal shelves have a more typical appearance, although there appears to be a delay in elevation. The tongue is typically seen as an obstructive influence on palatal elevation and fusion, typified by a Pierre Robin sequence in which micrognathia results in a relative macroglossia and elevation of the tongue, impeding palatal fusion [Tan and Farlie, 2013]. Analysis of snoopy mutants implies that the normal morphology of the palatal shelves and their successful elevation is dependent on the presence of the tongue to constrain growth and elongation of the shelves to the familiar sheet-like configuration and that this constraint is necessary for palatal fusion.
Recent molecular characterisation of ACS has led to the proposal that PLCB4 and GNAI3 mutations may result in a loss or reduction in endothelin signalling [Rieder et al., 2012; Clouthier et al., 2013; Gordon et al., 2013b]. Consistent with this hypothesis, mutations in EDN1, the gene encoding endothelin-1 itself, have also been identified in ACS and isolated the ‘question mark ear’ malformation [Gordon et al., 2013a]. Crkl has not been associated with the endothelin pathway previously, but given the significant phenotypic overlap between the snoopy mouse and ACS and the alteration to endothelin signalling in snoopy embryos, it is plausible that this is the case. Crkl is an adaptor protein involved in a number of kinase receptor signalling pathways that converge on Mapk/Erk signalling. Erk1/2 activation is a downstream consequence of signalling through a range of receptors including kinase receptors such as Fgfr and G protein-coupled receptors [Brown and Sacks, 2009]. The endothelin receptor A is a rhodopsin-like G protein-coupled receptor, and while Crkl has not been shown to interact with the endothelin receptor, it is possible that Crkl is involved in endothelin signalling, either directly or through transactivation via tyrosine kinase receptors. The snoopy phenotype is more extensive than that of the endothelin receptor A knockout [Ruest et al., 2004], and this likely reflects the role of Crkl in multiple growth factor and morphogen signalling systems.
The snoopy mouse models a number of key features of ACS, particularly with respect to the orofacial malformations. However, there are a number of features of the snoopy mouse that are not consistent with ACS, such as holoprosencephaly. The presence of holoprosencephaly is, however, common in the agnathia-otocephaly complex (AGOTC, OMIM 202650) [Guion-Almeida et al., 2002], an extremely rare and predominantly fatal condition involving agnathia, synotia, aglossia, microstomia, and holoprosencephaly, suggesting that snoopy may also recapitulate features of this condition. In addition, the description of ACS is based on postnatal features, and snoopy is limited to analysis at embryonic stages, since the interventions that have enabled ACS individuals to survive past the neonatal period, such as tracheostomy, are impractical in mice. We have not detected any temporo-condylar fusion in snoopy embryos, but this may be a reflection of the developmental stage at which the analysis was performed. The mandibular rudiment observed in severe mutants is very close to the skull base. If snoopy embryos were viable postnatally, the extended developmental timeframe may provide an opportunity for bony fusions to occur. Similarly, the ‘question mark ear’ malformation is a key feature in ACS. However, the external ears of mice are fundamentally different to those of humans and lack the distinct helix-lobe structure present in humans. It is, therefore, not possible to correlate the snoopy mice ears with the auricular phenotype of ACS.
The features in snoopy suggest that rather than being a candidate gene for ACS or AGOTC, CRKL functions in regulating the development of a series of tissues that overlaps with those affected in ACS and AGOTC. Alteration of retinoic acid signalling has been demonstrated in the Crkl null mouse [Guris et al., 2006], and Cyp26a1 expression is upregulated in snoopy embryos. Both the Crkl-Fgf8-Tbx1 and Edn-1 signalling networks are important for pharyngeal patterning. Thus, it is perhaps not so surprising that disruption of either signalling axis should produce overlapping phenotypes. AGOTC has been associated with heterozygous PRRX1 mutations [Sergi and Kamnasaran, 2011; Dasouki et al., 2013], and while there is no known interaction between Prrx1 and Crkl, endothelin-1 signalling may play a role in regulating Prrx1 expression [Doufexi and Mina, 2008]. The snoopy strain raises the interesting possibility that the retinoic acid and endothelin signalling systems not only co-exist, but also interact via Crkl to coordinate pharyngeal patterning. Thus, while additional work is required to clarify these relationships, based on phenotype and genetic interactions, it is possible that 22q11DS, ACS and AGOTC each result from a disruption of a molecular network in which CRKL is a common node.
Acknowledgements
This work was supported by the NHMRC grant No. 1002660 and the Victorian Government's Operational Infrastructure Support (OIS) Program. This work was enabled by the Australian Phenomics Network and in part supported by funding from the Australian Government's National Collaborative Research Infrastructure Strategy. T.Y.T. was funded by an Early Career Fellowship 607431 and J.F.B. is supported by a Senior Principal Research Fellowship, both from the National Health and Medical Research Council, Australia.
References
- 1.Andrews TD, Whittle B, Field MA, Balakishnan B, Zhang Y, et al. Massively parallel sequencing of the mouse exome to accurately identify rare, induced mutations: an immediate source for thousands of new mouse models. Open Biol. 2012;2:120061. doi: 10.1098/rsob.120061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Breckpot J, Thienpont B, Bauters M, Tranchevent LC, Gewillig M, et al. Congenital heart defects in a novel recurrent 22q11.2 deletion harboring the genes CRKL and MAPK1. Am J Med Genet A. 2012;158A:574–580. doi: 10.1002/ajmg.a.35217. [DOI] [PubMed] [Google Scholar]
- 3.Brown MD, Sacks DB. Protein scaffolds in MAP kinase signalling. Cell Signal. 2009;21:462–469. doi: 10.1016/j.cellsig.2008.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bruno DL, White SM, Ganesamoorthy D, Burgess T, Butler K, et al. Pathogenic aberrations revealed exclusively by single nucleotide polymorphism (SNP) genotyping data in 5000 samples tested by molecular karyotyping. J Med Genet. 2011;48:831–839. doi: 10.1136/jmedgenet-2011-100372. [DOI] [PubMed] [Google Scholar]
- 5.Caruana G, Farlie PG, Hart AH, Bagheri-Fam S, Wallace MJ, et al. Genome-wide ENU mutagenesis in combination with high density SNP analysis and exome sequencing provides a rapid route to the identification of novel mouse models of developmental disease. PLoS One. 2013;8:e55429. doi: 10.1371/journal.pone.0055429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Clouthier DE, Passos-Bueno MR, Tavares AL, Lyonnet S, Amiel J, Gordon CT. Understanding the basis of auriculocondylar syndrome: insights from human, mouse and zebrafish genetic studies. Am J Med Genet C Semin Med Genet. 2013;163:306–317. doi: 10.1002/ajmg.c.31376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Conti E, Grifone N, Sarkozy A, Tandoi C, Marino B, et al. DiGeorge subtypes of nonsyndromic conotruncal defects: evidence against a major role of TBX1 gene. Eur J Hum Genet. 2003;11:349–351. doi: 10.1038/sj.ejhg.5200956. [DOI] [PubMed] [Google Scholar]
- 8.Dasouki M, Andrews B, Parimi P, Kamnasaran D. Recurrent agnathia-otocephaly caused by DNA replication slippage in PRRX1. Am J Med Genet A. 2013;161:803–808. doi: 10.1002/ajmg.a.35879. [DOI] [PubMed] [Google Scholar]
- 9.Doufexi AE, Mina M. Signaling pathways regulating the expression of Prx1 and Prx2 in the chick mandibular mesenchyme. Dev Dyn. 2008;237:3115–3127. doi: 10.1002/dvdy.21762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fernández L, Nevado J, Santos F, Heine-Suñer D, Martinez-Glez V, et al. A deletion and a duplication in distal 22q11.2 deletion syndrome region. Clinical implications and review. BMC Med Genet. 2009;10:48. doi: 10.1186/1471-2350-10-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Garcia-Miñaur S, Fantes J, Murray RS, Porteous ME, Strain L, et al. A novel atypical 22q11.2 distal deletion in father and son. J Med Genet. 2002;39:E62. doi: 10.1136/jmg.39.10.e62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gong W, Gottlieb S, Collins J, Blescia A, Dietz H, et al. Mutation analysis of TBX1 in non-deleted patients with features of DGS/VCFS or isolated cardiovascular defects. J Med Genet. 2001;38:E45. doi: 10.1136/jmg.38.12.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gordon CT, Petit F, Kroisel PM, Jakobsen L, Zechi-Ceide RM, et al. Mutations in endothelin 1 cause recessive auriculocondylar syndrome and dominant isolated question-mark ears. Am J Hum Genet. 93:1118–1125. doi: 10.1016/j.ajhg.2013.10.023. (2013a). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gordon CT, Vuillot A, Marlin S, Gerkes E, Henderson A, et al. Heterogeneity of mutational mechanisms and modes of inheritance in auriculocondylar syndrome. J Med Genet. 50:174–186. doi: 10.1136/jmedgenet-2012-101331. (2013b). [DOI] [PubMed] [Google Scholar]
- 15.Guion-Almeida ML, Zechi-Ceide RM, Vendramini S, Kokitsu-Nakata NM. Auriculo-condylar syndrome: additional patients. Am J Med Genet. 2002;112:209–214. doi: 10.1002/ajmg.10631. [DOI] [PubMed] [Google Scholar]
- 16.Guris DL, Duester G, Papaioannou VE, Imamoto A. Dose-dependent interaction of Tbx1 and Crkl and locally aberrant RA signaling in a model of del22q11 syndrome. Dev Cell. 2006;10:81–92. doi: 10.1016/j.devcel.2005.12.002. [DOI] [PubMed] [Google Scholar]
- 17.Guris DL, Fantes J, Tara D, Druker BJ, Imamoto A. Mice lacking the homologue of the human 22q11.2 gene CRKL phenocopy neurocristopathies of DiGeorge syndrome. Nat Genet. 2001;27:293–298. doi: 10.1038/85855. [DOI] [PubMed] [Google Scholar]
- 18.Kurahashi H, Tsuda E, Kohama R, Nakayama T, Masuno M, et al. Another critical region for deletion of 22q11: a study of 100 patients. Am J Med Genet. 1997;72:180–185. doi: 10.1002/(sici)1096-8628(19971017)72:2<180::aid-ajmg10>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- 19.Lv S, Qin J, Yi R, Coreman M, Shi R, et al. CrkL efficiently mediates cell proliferation, migration, and invasion induced by TGF-β pathway in glioblastoma. J Mol Neurosci. 2013;51:1046–1051. doi: 10.1007/s12031-013-0096-3. [DOI] [PubMed] [Google Scholar]
- 20.Miller KA, Ah-Cann CJ, Welfare MF, Tan TY, Pope K, et al. Cauli: A mouse strain with an Ift140 mutation that results in a skeletal ciliopathy modelling Jeune syndrome. PLoS Genet. 9:e1003746. doi: 10.1371/journal.pgen.1003746. (2013a). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miller KA, Gordon CT, Welfare MF, Caruana G, Bertram JF, et al. bfb a novel ENU-induced blebs mutant resulting from a missense mutation in Fras1. PLoS One. 2013b;8:e76342. doi: 10.1371/journal.pone.0076342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moon AM, Guris DL, Seo JH, Li L, Hammond J, et al. Crkl deficiency disrupts Fgf8 signaling in a mouse model of 22q11 deletion syndromes. Dev Cell. 2006;10:71–80. doi: 10.1016/j.devcel.2005.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ogilvie CM, Ahn JW, Mann K, Roberts RG, Flinter F. A novel deletion in proximal 22q associated with cardiac septal defects and microcephaly: a case report. Mol Cytogenet. 2009;2:9. doi: 10.1186/1755-8166-2-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Padmanabhan RA, Nirmala L, Murali M, Laloraya M. CrkL is a co-activator of estrogen receptor α that enhances tumorigenic potential in cancer. Mol Endocrinol. 2011;25:1499–1512. doi: 10.1210/me.2011-0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rauch A, Zink S, Zweier C, Thiel CT, Koch A, et al. Systematic assessment of atypical deletions reveals genotype-phenotype correlation in 22q11.2. J Med Genet. 2005;42:871–876. doi: 10.1136/jmg.2004.030619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rieder MJ, Green GE, Park SS, Stamper BD, Gordon CT, et al. A human homeotic transformation resulting from mutations in PLCB4 and GNAI3 causes auriculocondylar syndrome. Am J Hum Genet. 2012;90:907–914. doi: 10.1016/j.ajhg.2012.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ruest LB, Xiang X, Lim KC, Levi G, Clouthier DE. Endothelin-A receptor-dependent and -independent signaling pathways in establishing mandibular identity. Development. 2004;131:4413–4423. doi: 10.1242/dev.01291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sergi C, Kamnasaran D. PRRX1 is mutated in a fetus with agnathia-otocephaly. Clin Genet. 2011;79:293–295. doi: 10.1111/j.1399-0004.2010.01531.x. [DOI] [PubMed] [Google Scholar]
- 29.Tan TY, Farlie PG. Rare syndromes of the head and face-Pierre Robin sequence. Wiley Interdiscip Rev Dev Biol. 2013;2:369–377. doi: 10.1002/wdev.69. [DOI] [PubMed] [Google Scholar]
- 30.Tan TY, Gordon CT, Amor DJ, Farlie PG. Developmental perspectives on copy number abnormalities of the 22q11.2 region. Clin Genet. 2010;78:201–218. doi: 10.1111/j.1399-0004.2010.01456.x. [DOI] [PubMed] [Google Scholar]
- 31.Verhagen JMA, Diderich KE, Oudesluijs G, Mancini GM, Eggink AJ, et al. Phenotypic variability of atypical 22q11.2 deletions not including TBX1. Am J Med Genet A. 2012;158A:2412–2420. doi: 10.1002/ajmg.a.35517. [DOI] [PubMed] [Google Scholar]
- 32.Williams LH, Miller KA, Dahl HH, Manji SS. Characterization of a novel ENU-generated myosin VI mutant mouse strain with congenital deafness and vestibular dysfunction. Hear Res. 2013;299:53–62. doi: 10.1016/j.heares.2013.02.006. [DOI] [PubMed] [Google Scholar]
- 33.Wurdak H, Ittner LM, Lang KS, Leveen P, Suter U, et al. Inactivation of TGFβ signaling in neural crest stem cells leads to multiple defects reminiscent of DiGeorge syndrome. Genes Dev. 2005;19:530–535. doi: 10.1101/gad.317405. [DOI] [PMC free article] [PubMed] [Google Scholar]