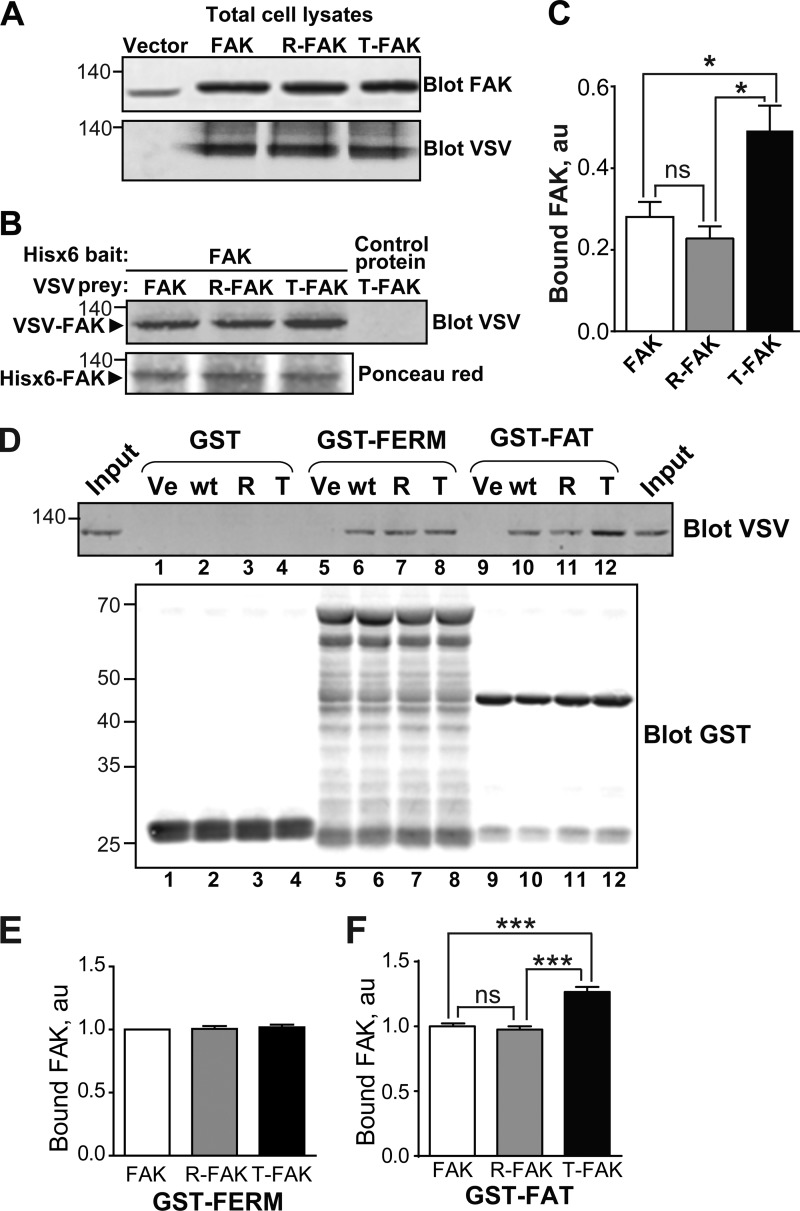
FIGURE 3.
T-FAT mutation increases full-length FAK dimerization and interaction with FAT. A–C, mutant and WT FAK proteins with N-terminal VSV tags were expressed by transfection in COS7 cells. A, immunoblotting of FAK in 20% of the amount of cell lysates used for each pull-down. Molecular mass marker positions are in kDa. B, cell lysates were loaded onto Ni2+ beads coated with recombinant His6-FAK (left lanes) or a His6-tagged 65-kDa unrelated protein (right lane). After extensive washing, bound proteins were eluted and the presence of VSV-tagged protein was assessed by immunoblotting. Ponceau red staining of the membrane shows equivalent amounts of His6-FAK bait. C, quantification of the pull-down experiment performed in triplicate (mean ± S.E., au, arbitrary units). One-way ANOVA, F(2,6) = 9.8, p = 0.013, Tukey's test; *, p < 0.05; ns, not significant. In these experiments, full-length FAK interactions are likely to involve FERM-FERM or FAT-FERM interactions, in addition to the FAT-FAT interactions. D, COS7 cell lysates containing identical amounts of VSV-tagged mutant or WT FAK proteins were added to beads coated with GST (lanes 1–4), GST-FERM (lanes 5–8), and GST-FAT (lanes 9–12). After washing and elution, bound FAK was visualized by immunoblotting with VSV antibodies. Equivalent quantities of the three different baits used (GST, GST-FERM, and GST-FAT) were revealed by immunoblotting with a GST antibody. Input (20% of the lysate amount used in the pull-down) is shown on either side of the VSV blot. E and F, quantification of the amount of VSV-FAK retained by GST-FERM (B) and GST-FAT (C) in three independent experiments (mean ± S.E., au, arbitrary units). One-way ANOVA GST-FERM, no significant difference, GST-FAT, F(2,6) = 43, p < 0.001, Tukey's test; ***, p < 0.001; ns, not significant. The T mutation in full-length FAK did not modify its binding to GST-FERM but increased its interaction with GST-FAT. A, B, and D, molecular mass marker positions are indicated in kDa.