FIGURE 2.
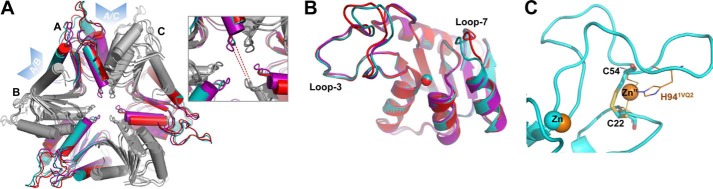
Overall structure of S-TIM5-dCD. A, superposition of the in crystallo hexameric forms (carried out in Coot). For clarity, every second protomer is shaded gray (S-TIM5-A in purple, S-TIM5-T in red, and S-TIM5-C in cyan). Blue arrowheads highlight interfaces between protomers. Inset, close-up of the middle region of the hexamer highlighting the greater compactness of the unbound S-TIM5-A structure (opposite protomers in the hexamer are ∼2.5 Å closer) as compared with the ligand-bound structures of S-TIM5-C and S-TIM5-T. B, superposition of the monomers from each structure. Zn2+ ions are displayed as spheres. C, secondary zinc binding motif in the S-TIM5-C (cyan) is substituted with a disulfide bond (represented by Cys22 and Cys54 as sticks). Secondary zinc (Zn”, orange sphere) and its binding residues (orange lines, His94 is labeled) from the structurally aligned T4 bacteriophage dCD (PDB code 1VQ2) are shown. Catalytic zinc ions are represented as overlapped cyan and orange spheres for S-TIM5-C and 1VQ2, respectively.
