Figure 1.
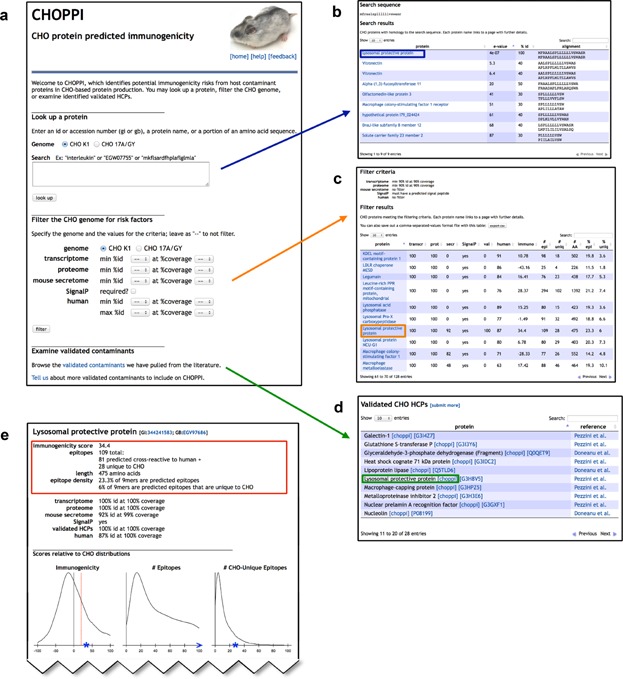
The CHOPPI web tool. CHOPPI allows two types of searches, “CHOP query” and “genome filter,” along with the ability to browse a set of validated CHOPs. These all lead into individual protein reports. (a → b) CHOP query. In this example, a fragment of an amino acid sequence is used as a query and CHO homologs to the query protein are identified. (a → c) Genome filter. In this example, the CHO genome is filtered for proteins with homology matches to both a CHO transcriptome and a CHO proteome, along with presence of a predicted signal peptide. The analysis of the filtered proteins is presented in a table. As with the CHOP query, an individual protein can then be examined. (a → d) The dataset of experimentally identified CHOPs. (one of {b, c, or d} → e) Individual protein report linked from the previous panel (boxed in each). The page summarizes both predicted epitope content (EpiMatrix whole-protein immunogenicity score, along with number and density of epitopes and CHO-unique epitopes), and predicted presence in protein production (homology matches against a CHO transcriptome, a CHO proteome, a mouse secretome, a set of validated CHOPs, and/or appearance of a predicted signal peptide). Immunogenicity evaluations are plotted relative to distributions over the whole genome.
