Figure 4. Biophysical characterisation of the library of G4 motifs.
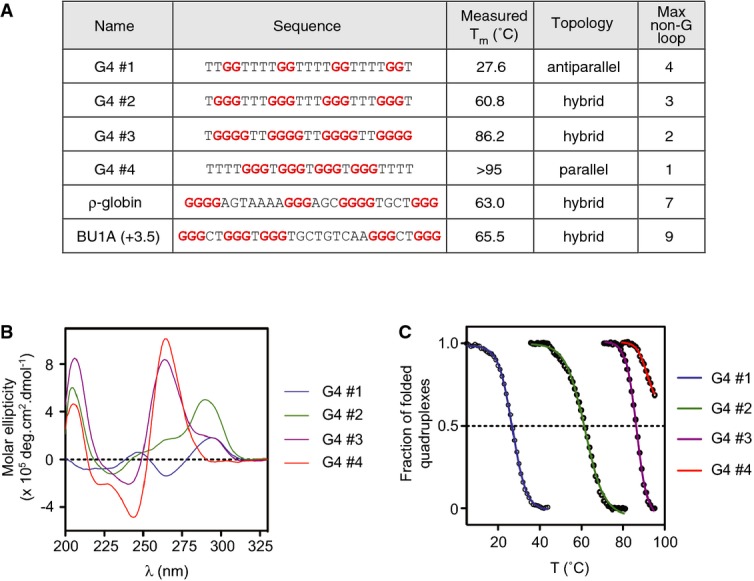
- Summary data table. G4#1–4 are model G4 motifs designed to have a range of melting temperatures. The in vitro topology was determined by circular dichroism in the presence of K+, shown in (B), and melting temperature is derived from the UV melting curves shown in (C).
