Abstract
Bacteria have evolved cellular control mechanisms to ensure proper length specification for surface-bound polysaccharides. Members of the Polysaccharide Copolymerase (PCP) family are central to this process. PCP-1 family members are anchored to the inner membrane through two transmembrane helices and contain a large periplasm-exposed domain. PCPs are known to form homooligomers but their exact stoichiometry is controversial in view of conflicting structural and biochemical data. Several prior investigations addressing this question indicated a nonameric, hexameric, or tetrameric organization of several PCP-1 family members. In this work, we gathered additional evidence that E.coli WzzB and WzzE PCPs form octameric homo-oligomeric complexes. Detergent-solubilized PCPs were purified to homogeneity and subjected to blue native gel analysis, which indicated the presence of a predominant high-molecular product of over 500 kDa in mass. Molecular mass of WzzE and WzzB-detergent oligomers was estimated to be 550 kDA by size-exclusion coupled to multiangle laser light scattering (SEC-MALLS). Oligomeric organization of purified WzzB and WzzE was further investigated by negative stain electron microscopy and by X-ray crystallography, respectively. Analysis of EM-derived molecular envelope of WzzB indicated that the full-length protein is composed of eight protomers. Crystal structure of LDAO-solubilized WzzE was solved to 6 Å resolutions and revealed its octameric subunit stoichiometry. In summary, we identified a possible biological unit utilized for the glycan chain length determination by two PCP-1 family members. This provides an important step toward further unraveling of the mechanistic basis of chain length control of the O-antigen and the enterobacterial common antigen.
Keywords: polysaccharide copolymerases, O-antigen elongation, electron microscopy, X-ray diffraction, low-resolution studies, oligomeric state
Introduction
Bacterial surfaces are covered by a diverse set of high-molecular weight sugar polymers that serve as a critical interface to interact either with host tissues or with the natural milieu of free living bacteria. Complex polysaccharide biosynthesis and export in gram-negative bacteria proceeds by several distinct mechanisms. The Wzy-dependent pathway relies on the inner-membrane resident glycosyl transferase (encoded in E. coli by the wzy gene), which catalyzes the polymerization of the oligosaccharide repeat units into a mature polymer within the periplasmic space.1,2 The biosynthesis of the repeat units of the polysaccharide takes place on the cytosolic face of the inner membrane where they are assembled on a lipid carrier, the C55 undecaprenol diphosphate. The entire lipid-oligosaccharide conjugate is transferred to the periplasm by a designated transporter (encoded in E. coli by the wzx gene) where it becomes the substrate for the aforementioned Wzy-polymerase that extends the chain.3 This mechanism of biosynthesis is utilized for the production of bacterial surface polysaccharides such as the lipopolysaccharide (LPS) O-antigen, the enterobacterial common antigen (ECA), and the bacterial capsules from Group 1 and 4.1
Another widespread mechanism of complex glycan synthesis relies on ABC-dependent transporters found within the inner membrane. The mature transporter is composed of two nucleotide binding domains and a cargo translocation channel formed by the transmembrane domains. The transmembrane portion and the ATP-hydrolyzing apparatus is encoded by two distinct genes, exemplified in E. coli by kpsT and kpsM genes.4,5 In this system, the entire glycan is synthesized in the cytosol by the action of specific glycosyl transferases, which transfer the sugar moiety from the activated donor to the nonreducing end of the growing polysaccharide chain. The two pathways are sometimes found to operate in parallel in certain bacterial species including Pseudomonas aeruginosa.6,7
The number of repeat units is not random and mature O-antigen and ECA polymers display a unimodal distribution of lengths. The distribution is imposed by the chain-length regulator proteins, which are indispensable for the proper function of both pathways, and are anchored at the inner membrane and designated as the Polysaccharide Copolymerases (PCP). Members of this large superfamily of bacterial proteins are present in a wide range of bacterial species and are involved in the export of a diverse set of glycans. PCPs exhibit a characteristic topology where a large periplasmic domain is anchored on each end to the inner-membrane by a transmembrane and/or aliphatic helix.8 Most PCPs also contain a proline-rich segment proximal to and overlapping the C-terminal trans-membrane helix.9
PCPs are classified into several different families. The PCP-1 family members are essential for biosynthesis of glycans making up the LPS O-antigen and ECA and are exemplified by E. coli WzzB and WzzE proteins, respectively. Both WzzB and WzzE operate as chain-length regulators for their respective polysaccharides by controlling the number of repeat units incorporated into the growing polysaccharide chain.10,11 Members of this subfamily contain only short cytoplasm-exposed regions at their N- and C-termini. PCP-2 family members contain a number of tyrosine residues within the cytosol-exposed C-terminus and undergo cycles of phosphorylation and dephosphorylation by their respective tyrosine kinases and tyrosine phosphatases.12 PCP-2A are found in Gram-negative bacteria and are involved in Wzy-dependent capsular polysaccharide export. The C-terminal, cytoplasmic kinase domain is responsible for tyrosine phosphorylation. PCP-2B are involved in the biosynthesis and export of the capsular polysaccharides in Gram-positive species.13 In this class the tyrosine kinase is expressed as a separate polypeptide. The exact topology of PCP-3 family members remains to be resolved. KpsE from E. coli K1 was proposed to contain only one transmembrane domain near the N-terminus and a membrane-associated at the C-terminus.14 On the other hand, CtrB from Neisseria meningitis was shown to contain both N- and C- terminal transmembrane domains15 much like PCP-1, and -2 family members.
Here we are interested in PCP-1 proteins. It is not completely clear whether or not formation of the proper oligomeric complex in vivo by WzzB and related molecules is essential for O-antigen chain length determination. However, some mutations that influenced the PCP function were shown to affect oligomerization.16,17 Other investigations were less clear about the correlation between PCP-1 oligomerization and very-long O-antigen chain-length determination.18 Establishing the subunit organization and stoichiometry of the chain-length regulators constitutes therefore an important step toward understanding their function. Initial attempts to establish the oligomeric state of the full-length PCP-1 proteins relied on chemical crosslinking but these experiments did not provide a clear answer on the oligomeric composition.19 The first low-resolution structural studies by small-angle X-ray scattering (SAXS) of the detergent-solubilized WzzB reported a tetrameric assembly as a likely oligomeric state of the protein.20 The first high-resolution crystal structures of the periplasmic domain of three PCP-1 family members, WzzB, WzzE, and FepE, demonstrated directly that these proteins assemble into oligomers.21 In the crystals, the three proteins adopted bell-shaped structures with distinctly different oligomeric states, WzzB being a pentamer, WzzE an octamer and FepE a nonamer.21 A follow-up study on the chimeric and mutant PCP-1 family members revealed the ability of the WzzB periplasmic domain to assemble into octamers.22 An independent electron microscopy analysis of the 2D crystals of the full-length PCP-1 proteins suggested; however, a hexameric arrangement of the protomers for the PCPs aforementioned.23 In situ crosslinking of FepE also suggested hexameric arrangement.18 Moreover, two studies examining the protein complexes found in E. coli K-12 membranes using blue native PAGE reported distinctly different oligomeric states for WzzB. While one report indicated WzzB to be a dimer,24 the other study reported oligomers with molecular weights more than 500 kDa.25
In light of the above experimental observations, we continued investigations of the full-length PCPs, WzzB, and WzzE from E. coli, in order to resolve the nature of their quaternary organization. We characterized these detergent-solubilized PCPs biochemically by size exclusion chromatography (SEC) coupled with multiangle laser light scattering, and structurally by negative staining electron microscopy, electron tomography, and low resolution X-ray crystallographic analysis. Our experimental results provide further strong affirmation of the octameric nature of the homo-oligomeric complexes formed by full-length WzzB and WzzE.
Results and Discussion
We have previously determined the crystal structure of the periplasmic domain of WzzE in an bell-shaped octameric form and showed from single particle reconstruction by negative stain electron microcopy that the full-length protein assembles into particles of shape and size corresponding very well to those determined from the crystal structure.21 The presence of the transmembrane helices stabilized the oligomeric structure. On the other hand, the periplasmic domain of WzzB was crystallized in several oligomeric forms, as bell-shaped octamers, vase-like pentamers open at the top, and open trimers.21,22 Of these, the octamers showed the most intimate protomer–protomer contacts, suggesting this to be the most stable oligomer. To further investigate the role of the transmembrane helices on the oligomeric structure of WzzB as well as WzzE, and to resolve the long-standing ambiguity surrounding the subunit stoichiometry of WzzB from E. coli K-12 and WzzE from E. coli O7:H157 we have pursued biophysical and structural investigations of the full-length PCPs.
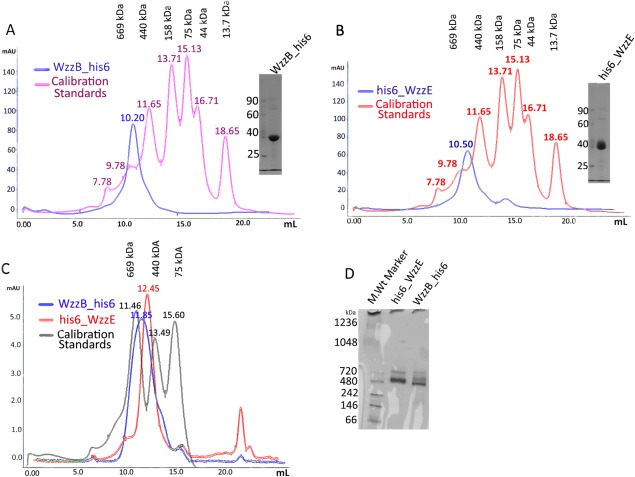
The full-length WzzB and WzzE proteins were overproduced in E. coli membranes and initially extracted with DDM. To assess the effect of various detergents on the apparent oligomeric state of WzzB and WzzE, the DDM was exchanged with other detergents on Co-NTA affinity resin and the protein was subjected to SEC. Commonly utilized detergents, such as decylmaltoside (DM), β-octylglucoside (OG), and n-Dodecyl-N,N-dimethylamine-N-Oxide (LDAO) were among those tested (Table S1, Supporting Information). The SEC profile indicated that some detergents supported the oligomeric assembly while others disrupted oligomers or denatured the protein (Fig. S1, Supporting Information); their effects were protein specific. WzzB could be purified in nonyl-glucoside producing a sharp symmetric peak whereas WzzE tended to precipitate in the same detergent (data not shown). Sodium N-lauryl sarcosine was useful for membrane solubilization of WzzE whereas WzzB did not tolerate this notoriously harsh detergent. The elution profile of β-DDM-extracted WzzE and WzzB from the Superdex-200 size exclusion column, calibrated with soluble globular protein standards, indicated that both proteins assembled into very high molecular mass oligomers of over 500 kDa in size [ Fig. 1(A,B)]. This suggested that both proteins maintained a supramolecular assembly following the solubilization by this detergent. In sharp contrast, their corresponding periplasmic domains formed much smaller oligomers in solution at low concentration (∼2 mg/ml), most likely dimers, but assembled into oligomers during the crystallization process at high protein concentration.21,22 Furthermore, WzzE and WzzB eluted at a similar retention volume of about 10.5 and 10.2 mL, respectively, suggesting that both oligomeric complexes are composed of the same number of subunits [ Fig. 1(A,B)]. When a Sepharose-6 column with a wider separation range was used, the two protein-detergent conjugates eluted further apart (11.85 mL for WzzB vs. 12.45 mL for WzzE) but also fell within 440 and 669 kDa standards as before [ Fig. 1(C)].
Figure 1.
Purification and preliminary native gel-based analysis of WzzB and WzzE oligomers. Size-exclusion elution profiles of (a) WzzB and (b) WzzE purified on Superdex-200 column in β-dodecyl-maltoside (β-DDM) as well as the Coomassie-blue stained SDS-polyacrylamide gels demonstrating the purity of each protein after the size-exclusion step; (c) Size-exclusion chromatography of WzzB-β-DDM and WzzE-β-DDM analyzed on Superose-600 column. The SEC trace of standard globular proteins is shown in gray and corresponds to the following molecular masses: 660 kDa—Thyroglobulin, 440 kDa—Apo Ferritin, 158 kDa—Aldolase; (d) Blue native PAGE analysis of the WzzB-β-DDM and WzzE-β-DDM oligomers.
We further analyzed both proteins by blue native PAGE (BN-PAGE) [ Fig. 1(D)]. The Coomassie Blue dye imparts a uniform negative charge distribution onto proteins resulting in their separation mainly based on the molecular weight. Indeed, the BN-PAGE analysis of both WzzB and WzzE produced very similar electrophoretic migration patterns with the predominant bands corresponding to molecular masses between the 480 and 720 kDa standards for each protein [ Fig. 1(D)]. This further supported the notion that both WzzE and WzzB form oligomers of similar stoichiometric composition. However, the resolving power of BN-PAGE under our experimental conditions was insufficient for accurate determination of the molecular weight of each PCP assembly.
The protein complexes contain an unknown amount of bound β-DDM molecules. We combined size-exclusion chromatography with multi-angle laser light scattering (SEC-MALLS) and refractive index measurements to better estimate the molecular mass of the oligomeric assemblies formed by each PCP (Fig. 2). SEC-MALLS analysis estimated the molecular mass for each protein-β-DDM complex at 500 kDa for WzzE (±20 kDa) and 550 kDa for WzzB (±10 kDa). Once again, in line with the previous observations, SEC-MALS analysis demonstrated that the oligomeric assemblies formed by both proteins were of very similar molecular weight suggesting the same quaternary organization. The molecular mass of WzzB is 36 kDa while WzzE is 40 kDa. With an unknown amount of detergent bound to the oligomer, these data alone indicate that the number of protomers exceeds eight unless the β-DDM contributes as much as 50% of the molecular weight of the protein-DDM complex.
Figure 2.
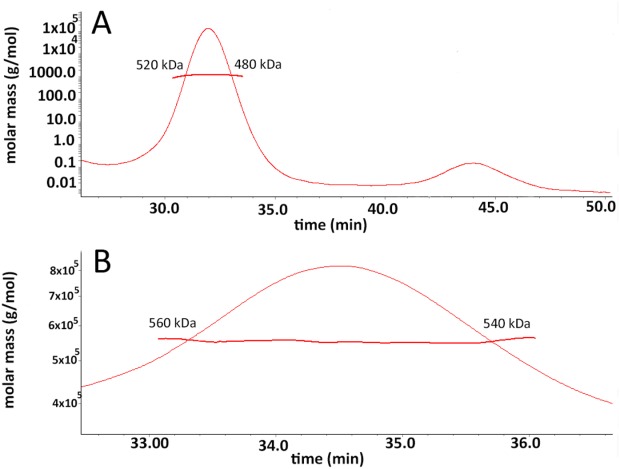
SEC-MALS analysis of β-dodecyl-maltoside solubilized WzzB and WzzE. (a) The raw size exclusion trace of WzzE injected onto Superdex-200 column with the curves corresponding to A280 (green), light scattering (red), and refractive index (blue) overlaid; (a) Calculated molecular mass of the peak and the variation in the molecular weight across the peak for β-DDM WzzE; and (b) Molecular mass of the peak and the variation across the β-DDM WzzB as calculated by Astra™ computational software package.
To obtain more direct information on the subunit stoichiometry, we applied electron microscopy and X-ray diffraction methods. For these studies WzzB was solubilized with β-DDM while WzzE was extracted from the membranes using sodium N-lauryl sarcosine and exchanged into one of the detergents outlined in Table S1, Supporting Information. The final yield of each protein was ∼0.7 mg/L. We were able to obtain crystals of both proteins. However, crystals of WzzB diffracted only to ∼20 Å resolution despite extensive detergent screening and optimization. Space group and unit cell dimensions could not be extracted from the diffraction images of these crystals. WzzE purified in the presence of LDAO formed better diffracting crystals, which after optimization diffracted to 6 Å resolution (Table I).
Table I.
Data Processing and Refinement Statistics for WzzE-LDAO
| Data collection | |
|---|---|
| Synchrotron beamline | 23ID-B at APS |
| a, b, c (Å); β(°)a | 276.8, 246.3, 133.2; 89.98 |
| wavelength(Å) | 0.97899 |
| resolution (Å)b | 50–6.00 (6.1–6.0) |
| observed hkl | 43,396 |
| unique hkl | 12,398 |
| redundancyb | 3.5 (2.8) |
| completeness (%)b | 97.2 (90.8) |
| Rsym | 8.4% |
| I/(σI)b | 14.9 (0.9) |
| Wilson B (Å2) | 258 |
| Refinement | |
| Twin operator | (-h, -k, l) |
| Twin ratio | 0.255 |
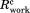 (# hkl) (# hkl) |
0.23 |
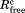 (# hkl) (# hkl) |
0.255 |
| Ramachandran plot | |
| Allowed (%) | 98.8 |
| Generous (%) | 1.0 |
| Disallowed (%) | 0.2 |
| RMSD | |
| Bonds (Å) | 0.012 |
| Angles (°) | 2.25 |
| PDB code | 4WL1 |
Pseudo orthorhombic Bravais lattice. Data reduction in orthorhombic space group gives very high Rsym.
Data for the highest resolution shell are given in parentheses. Rwork = (∑Fobs − Fcalc|)/∑Fobs.
cRsym = (∑|Iobs − Iavg|)/∑Iavg.
Since we have previously determined the structure of the periplasmic domain of WzzE at 2.4 Å resolution, we reasoned that at 6 Å resolution we should be able to determine the oligomeric state of WzzE in the crystal utilizing molecular replacement methods. The periplasmic domain of WzzE alone formed octamers in the crystal and our previous negative stain electron microscopy analysis of WzzE showed good correlation with the crystal structure of its periplasmic domain.21 However, this oligomeric state has since been questioned based on the EM analysis of 2D crystals, which were interpreted in terms of a hexameric WzzE.23
A patterson self-rotation function was calculated to obtain an inference on the possible organization of the asymmetric unit. Two peaks corresponding to an 8-fold noncrystallographic (NCS) symmetry axis (κ = 45° section) were found to lie at φ = 32.5, ψ = 47.5, and φ = 147.5 ψ = 132.5 (Fig. 3). Moreover, four peaks corresponding to a 4-fold noncrystallographic symmetry (κ = 90°) were identified at φ = 32.5. ψ = 47.5; φ = 147.5. ψ = 132.5; φ = 32.5, ψ = 130.5; φ = 147.5, ψ = 50.0 (Fig. 3). No strong peaks were seen at kappa sections κ = 120, 72° or 60° corresponding to three-, five-, or six-fold noncrystallographic symmetry. This analysis supported the presence of octameric oligomers. Nevertheless, various oligomeric forms of the periplasmic domains of WzzE, FepE and WzzB proteins21 were utilized as search models in the molecular replacement method. A convincing solution was obtained with the periplasmic domain of WzzE (PDB code 3B8O), giving four octamers in the asymmetric unit [ Fig. 4(A)]. The refinement led to Rwork of 0.23 and Rfree of 0.26 and gave us confidence in this solution. Inspection of the arrangement of molecules in the crystal lattice showed reasonable packing, with large contact areas between the periplasmic domains [ Fig. 4(A,B)]. A second set of crystal contacts is formed by the arrangement of octamers in the same bottom-to-bottom configuration. This arrangement of the molecules correlates well with the output of the self-rotation function. Strong peaks seen at kappa section = 45° (φ = 32.5, ψ = 47.5 and φ = 147.5, ψ = 132.5) correspond to 8-fold (NCS) along the direction of the central axis of each pair of octamers found in an end-to-end configuration. The same polar angles of two of the four-fold NCS peaks represent four-fold symmetry of the octamers. The other 2 four-fold NCS relate the independent octamers [ Fig. 4(A,B)], which corroborates the self-rotation function with the derived final model.
Figure 3.
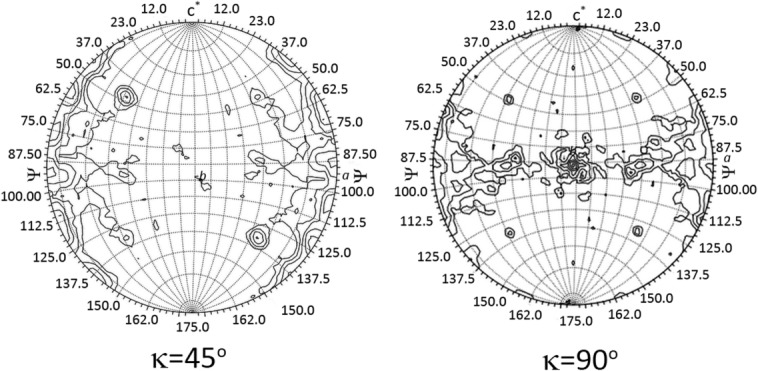
Kappa sections 45 and 90 of the Patterson self-rotation function corresponding to 8-fold and 4-fold noncrystallographic symmetry, respectively; [Phi (Φ) angles are denoted by lines running from top to bottom whereas Psi (ϕ) angles are running from left to right]. Kappa sections were generated with the program GLRF26 using 30 Å radius of integration and 6–12 Å data.
Figure 4.
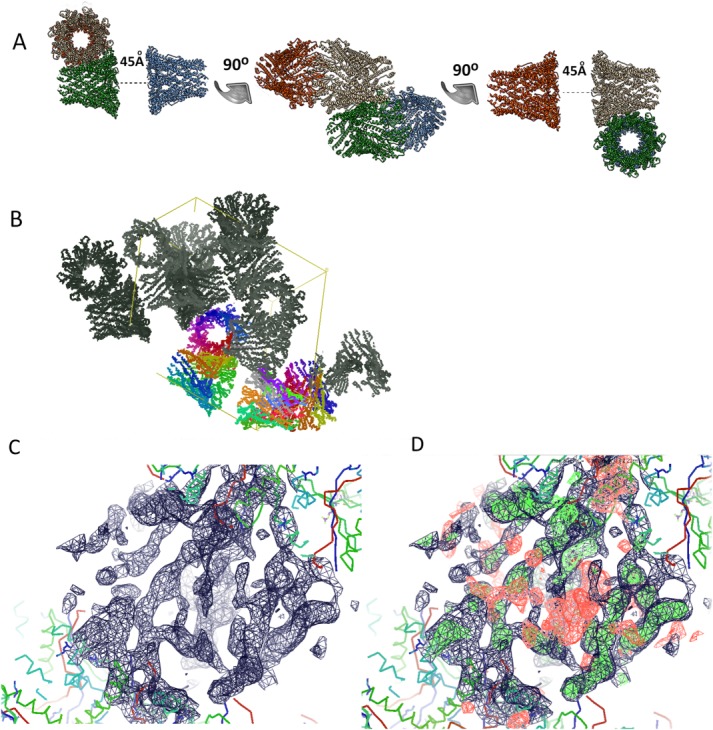
Crystallographic analysis of WzzE. (a) The 6 Å crystal structure with four octamers in the asymmetric unit. The distance between the periplasmic domains of the octamers is shown. The figure was prepared using the UCSF Chimera; (b) A crystallographic unit cell showing the packing of the molecules into the crystal lattice; (c) 2Fo-Fc (contoured at 1σ) and (d) 2Fo-Fc overlaid on Fo-Fc (contoured at 2.2σ) electron density maps for the missing part of the model in interperiplasmic domain regions.
The two interacting WzzE oligomers form extensive interactions between their cytosolic and transmembrane domains with the periplasmic domains 45 Å apart. This dumbbell arrangement and distance between the periplasmic domains is similar to the previous EM reconstruction of WzzE in negative stain EM analysis.21 Such an arrangement decreases exposed hydrophobic surface and decreases the number of the detergent molecules around TM helices. Given the short distance between the periplasmic domains, it is very likely that the conformation of the TM helices in this arrangement is different from what is normally expected in the cell membrane. We surmise that the difficulties we encountered to improve the diffraction of the crystals are in fact largely due to the nonuniform conformations adopted by the TM domains while the periplasmic domains exhibit very tight packing [ Fig. 4(B)]. The electron density in the region between the periplasmic domains [ Fig. 4(C,D)] is insufficient to model either the detailed architecture of the TM helices or the short cytosol-exposed regions. In summary, the crystallographic analysis of WzzE combined with the previously determined EM negative stain reconstruction and the octameric structure of the periplasmic domains21 firmly establishes the octameric nature of the full-length WzzE protein. The bottom-to-bottom arrangement of the two octamers in the crystals is mediated by the interactions between transmembrane helices of the two oligomers and does not bear physiological relevance.
To investigate the oligomeric state of WzzB and in view of the poorly diffracting crystals we turned to negative stain electron microscopy analysis. Examination of the grids revealed the presence of several distinct types of particles in the WzzB purified in DDM. The small, round particles correspond to the top view, while the medium particles to a side view of single WzzB oligomers [WzzB-1 and WzzB-2, respectively, Fig. 5(A)]. The larger particles show two WzzB oligomers associated bottom-to-bottom through their cytosolic and transmembrane parts [referred to as 2X WzzB, Fig. 5(A)]. The latter view is similar to that previously observed for WzzE by negative stain electron microscopy,21 except that the distance between the periplasmic domains of the oligomers is much larger, ∼120 Å. Image classification of the smaller particles did not lead to a clear set of class averages and therefore they were not analyzed further. Approximately 1,000 larger particles were chosen for the classification [ Fig. 5(B)] and produced a very distinct class average [ Fig. 5(C)], from which the dimensions of the oligomers were measured using the UCSF Chimera software.27 Each octamer was found to measure 150 Å in length, 60 Å across the top and 110 Å across the bottom. The periplasmic domain of this class average projection was measured to be 100 Å in length. These dimensions of the periplasmic segment are in good agreement with those derived from a crystal structure of the octameric periplasmic domain of WzzB (100 Å in length, 50 Å across the top, and 90 Å across the bottom)22 and the crystal structure superposed very well with the EM projection map [ Fig. 6(C)].
Figure 5.
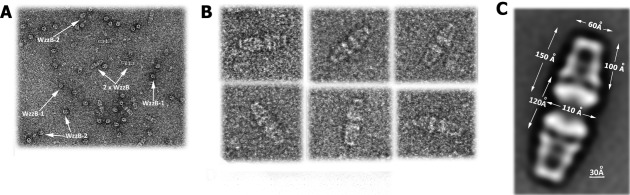
Negative stain electron-microscopy analysis of β-dodecyl-maltoside solubilized WzzB. (a) A snapshot of the EM grid area demonstrating the two populations of particles formed by WzzB as observed at 68,000× magnification; (b) A gallery of the images of the representative larger particles used for the class average calculations; (c) A projection of the class average based on 1,000 particles of a larger assembly with the calculated dimensions labeled in white.
Figure 6.
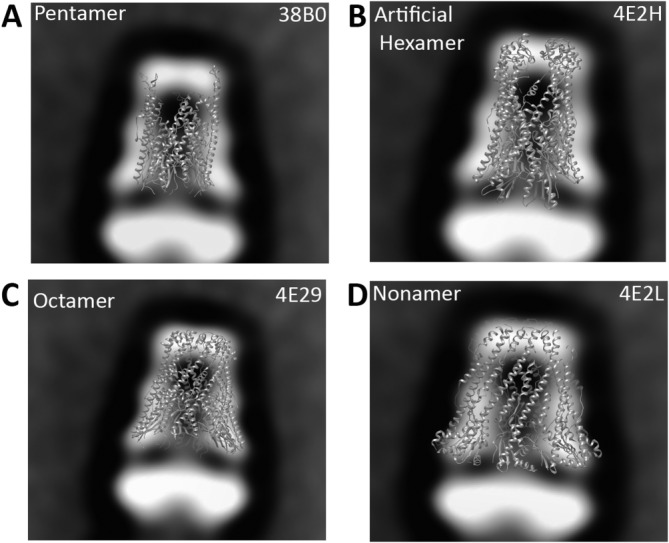
Crystal structures of the previously determined periplasmic domains of WzzB (a–c) and WzzBFepE (d) superposed with the EM projection of 2× WzzB class average. PDB code of each X-ray structure used is specified in the top right corner.
One notable difference is the diameter across the top (50 Å in the crystal vs. 60 Å in the EM). This is likely due to random orientations adopted by flexible loop regions, which were found to cap the oligomer in in the X-ray structure of the chimeric WzzB proteins.22 In the negative stain analysis they may contribute to the EM reconstruction resulting in a slightly bigger diameter across the top of the molecule. The dimensions derived from the negative stain EM analysis are also comparable to those obtained by Larue et al. for liposome-resuspended WzzB from S. typhimurium (100 Å across the bottom, 55 Å across the top, and 95 Å in length) notwithstanding a different interpretation of the oligomeric state by these authors.23 The detergent belt around the trans-membrane regions of the protein will contribute to the observed projection since the heavy atom salt used in negative staining is not expected to penetrate the detergent layer.28 However, the dimensions of the periplasmic segments should not be influenced by the presence of the detergent. The structure also demonstrates that the transmembrane domains and the cytosolic regions of WzzB extend for ∼60 Å below the periplasm. We tested the fit of oligomers with different number of protomers to the EM-derived envelope. These included the pentamer of the periplasmic domain of WzzB from S. typhimurium [PDB code 3B8P, Fig 6(A)], an artificial hexamer composed of two trimers of WzzBSF [PDB code 4E2H, Fig. 6(B)], the octamer of the chimeric S. flexneri–S. typhimurium WzzB [PDB code 4E29, Fig. 6(C)], and the nonamer of WzzFepE [PDB code: 4E2L, Fig. 6(D)]. Clearly, the best fit is for the octamer. These results strongly support the notion that the most stable form of full-length WzzB is indeed an octamer. The distance of ∼120 Å between the periplasmic regions in WzzB particles [ Fig. 5(C)] is much larger than observed in the WzzE particles by EM reconstruction.21 This large distance suggests that the two TM bundles interact through their cytoplasmic ends and do not intertwine. Several explanations can be offered to rationalize this difference. First of all, the different arrangement may be attributed to a different chemistry of the detergents used for the two analyses. The negative stain EM analysis of WzzB was carried out in the presence of neutral β-DDM while the WzzE preparations for crystallization and EM contained a zwitterionic detergent LDAO. Second, the difference in the TM helix arrangement may be a reflection of amino acid difference within the TM regions; the TM helices of WzzE and WzzB differ in ∼50% of amino acids.
The presence of bell-shaped octamers and bell hexadecamers suggests the interpretation of SEC-determined molecular weights. The observed size exclusion peak [ Fig. 1(A,B)] may arise from the equilibrium in solution between single octamers and the dumb-bell hexadecamers interacting bottom-to-bottom, much like the orientation observed in the negatively stained EM and in the crystals. Many integral monotopic membrane proteins are known to oligomerize through their hydrophobic membrane-anchored helices when solubilized in detergent.29,30 Sixteen copies of WzzE and WzzB would have molecular weights of 632 and 576 kDa, respectively, The octamers would have half that mass. We favor the presence of a dynamic equilibrium between octamers and hexadecamers resulting in broad SEC peaks with intermediate molecular weight maxima.
Conclusions
Although current investigations do not directly lead to an understanding of the mechanism of the glycan chain-length regulation performed by either protein, it offers new insights into the long-standing question of their subunit stoichiometry. The ability of the chain-length regulators to form higher order homo-oligomeric complexes is essential for chain length regulation16,17,19 and some experimental evidence suggests that the growing polysaccharide may be interacting directly with the periplasmic portion of the PCP-1 proteins.20,31,32 The number of subunits forming an oligomer may thus be directly linked to the length imposed on the growing glycan polymer, as it would influence the overall surface area available for the polysaccharide-protein interactions. Taken together, data presented here show that the octameric arrangement of the protomers is compatible with all available data and is the most likely oligomeric form that exists in the bacterial inner membrane. We also conclude that the pentameric structure observed in the crystal of S. typhimurium WzzB21 is an artifact of crystallization. While in all other oligomers the protomers interact along their entire length, the pentamers are formed through contacts of the base domain only, while the long helices diverge from the center of the oligomer. The nonamers formed by the periplasmic domain of WzzFepE under several different crystallization conditions21,22 may represent another oligomeric state for a subclass of PCP-1 and may be related to a very long chain length of the LPS O-antigen of over a 100 repeat units imparted by this PCP.
Material and Methods
Expression and purification
The gene encoding full-length WzzB protein from E. coli MGH1655 strain was PCR-amplified from the genomic DNA and ligated into the EcorI and KpnI sites of the arabinose-inducible pBAD vector, pBAD24 (AmpR). The C-terminal hexa-histidine tag was introduced during the PCR. Full length WzzE from E. coli O157:H7 EDL33 was subcloned from the genomic DNA using standard molecular biology techniques into the BamHI and EcoRI sites of the pET-based vector carrying a non-cleavable N-terminal hexa-His (pFO4, AmpR) tag.
The constructs were transformed into C43 (DE3) E. coli expression strain33 and small expression tests was performed by inducing 10 mL of culture grown in LB broth until the OD600 was 0.6–0.8. Several concentrations of isopropyl-β-d-1-thiogalactopyranoside (IPTG) or Arabinose were tested either O/N at 22°C or for 6 h at 30°C. The conditions found to result in the highest protein expression included induction at O.D. of 0.6 with 0.350 mM IPTG and 0.2% (w/v) L-(+)-Arabinose for WzzE and WzzB, respectively. The expression was allowed to proceed for 6 h at 30°C. The cells from 6 to 12 L of cultures grown in TB broth were harvested by spinning the cultures at 3000g. The cells were then resuspended in buffer containing 30 mM Tris pH 7.4, 200 mM NaCl, 10 mM imidazole, 5% glycerol, and 0.5 mM 3,3′,3″-Phosphanetriyltripropanoic acid (TCEP) at a ratio of 4 mL of buffer to 1 g of wet cell paste. Resuspended cells were subjected to two passes through the Avestine Emulsiflex C3 Cell Disruptor at ∼22 PSI. Remaining cells and cell debris were separated from the soluble fraction by spinning the lysate at 14,000g for 45 min. The membranes were isolated by ultra-high-speed centrifugation of the soluble fraction at 150,000g for 45 min. The collected membranes were subsequently resuspended in resuspension buffer in a ratio of 1 mL of buffer per 0.5 g of the isolated membrane pellet and either stored at −80°C for further use or solubilized immediately. WzzB was solubilized in 2% (w/v) n-dodecyl-β-d-maltoside (β-DDM, highest purity Anagrade™, Anatrace, CA) while WzzE was in 2% (w/v) sodium N-lauryl sarcosine (99% pure analytical grade, Sigma Aldrich) for 30 min at 4°C.
The insoluble material was separated by another ultra-high-speed spin at 150,000g for 30 min. The solubilized fraction was further diluted with the resuspension buffer to decrease the detergent concentration below 1%, and incubated overnight with 1.5 mL of TALON™ Cobalt affinity resin (Clonetech). The resin was loaded into a column and washed once with 25 mL of each of the following buffers containing the appropriate detergent at a concentration of 0.5 mM for β-DDM or 4 mM for n-dodecyl-N,N-Dimethylamine-N-Oxide (LDAO) (Wash buffer 1: 30 mM Tris pH 7.4, 200 mM NaCl, 10 mM imidazole, 5% (w/v) glycerol, and 0.5 mM (TCEP); Wash buffer 2: 30 mM Tris pH 7.4, 1.0M NaCl, 20 mM imidazole, 5% (w/v) glycerol, 0.5 mM TCEP; Wash buffer 3:30 mM Tris pH 7.4, 200 mM NaCl, 40 mM imidazole, 5% (w/v) glycerol, and 0.5 mM TCEP). The protein was eluted in an elution buffer containing 30 mM Tris pH 7.4, 200 mM NaCl, 200 mM imidazole, 5% (w/v) glycerol, and 0.5 mM TCEP. The protein purity was assessed on a 10% SDS–PAGE denaturing gel. The protein was subsequently concentrated using a 100 kDa molecular mass cut-off concentration filter unit (Amicon) and injected onto either Superdex-200 10/300 GL or Superose-6 10/300 GL size exclusion columns equilibrated with the elution buffer containing in addition 0.5 mM DDM (for WzzB) or 4 mM LDAO (for WzzE). The concentration of the injected protein varied between 1 and 4 mg/mL depending on the preparation. The protein containing fractions were pooled and concentrated to 3–5 mg/mL using a concentrator with a 100 kDa molecular mass cut-off.
Crystallization of WzzE and structure determination
WzzE was solubilized in sodium N-lauryl sarcosine. This detergent was exchanged into 20 different commercially available detergents on the TALON™ Co2+ affinity resin (Clonetech). Symmetrical, Gaussian-shaped peaks on a size exclusion column were seen in many different detergents and these preparations were subjected to the sparse-matrix screening using a Gryphon™ crystallization robot (Art Robbins Instruments) in a sitting drop vapor diffusion format using several commercially available and home-made crystallization screens. Successful crystallization hits were further reproduced and optimized in a hanging-drop format. The crystals had largest dimension of 50−70 μm and were screened for diffraction properties at the Canadian Light Source CMCF-ID beamline. The first WzzE crystals (purified in β-DDM) diffracted only to ∼20 Å. This prompted us to explore other detergents (Table S1, Supporting Information). WzzE crystallized in a number of conditions when solubilized in different detergents (Table S1, Supporting Information). Of the nearly ∼400 screened crystals most diffracted to between 20 and 12 Å. The most significant improvement in the resolution of the diffraction pattern was seen when WzzE was purified in 4 mM LDAO. The best crystals grew in the sitting drop set up in micro-bridges (Hampton Research) over a reservoir consisting of 8% PEG 8000, 0.1M Bis-Tris buffer pH 6.5, 0.2M NaCl, 15% glycerol with the drop containing1 μL of protein solution and 0.5 μL of reservoir. Micro seeding was used to obtain the best crystals. Because of the high glycerol concentration, no further cryo protectant was needed and the crystals were directly frozen in liquid nitrogen. Secondary detergent screening and various dehydration strategies led to no further improvements in diffraction quality. Ultimately, two complete datasets were collected on two different crystals from the same crystallization condition. The first data set was collected at the CMCF-ID beamline at the Canadian Light Source to 7 Å resolution, while the second one was collected at the 23ID beamline at the Advanced Photon Source to a resolution of 6 Å.
We were able to obtain crystals of WzzB solubilized and purified in DDM; however, despite much effort no crystals diffracting beyond ∼20 Å could be obtained. Neither spacegroup nor unit cell parameters could be assigned based on the limited diffraction data. These crystals were not pursued further.
The WzzE data were processed and scaled using HKL2000 suite.34 Although cell dimensions suggested an orthorhombic space group, the Rsym was 0.18. Of the three possible monoclinic settings only one had a low Rsym of 0.084 identifying the true crystallographic two-fold axis. Twinning analysis using the Xtriage program of the Phenix suite35 indicated the presence of a twin operator (-h,-k, and l) in both datasets. The proportion of twinning was 0.255 for the 6 Å and 0.311 for the 7 Å dataset. The structure was solved by molecular replacement. The various previously determined periplasmic domains of PCP oligomers with nine, eight, six, and five protomers per oligomer21 were used as search models using Phaser.36 The only reasonable solution was obtained with the periplasmic domain of WzzE (3B8O). Four octamers are present in the asymmetric unit. The refinement was performed with Phenix_Refine applying NCS restraints, secondary structure restraints, restraints to the periplasmic domain as a reference model and utilizing a group B-factor per residue. The twinning with a twin operator -h, -k, and l was utilized in the refinement. The model includes residues Glu55 to Asp319 of the periplasmic domain of WzzE with residues 86–94 and 231–236 missing. The final Rwork of 0.23 and Rfree of 0.25 afforded confidence in the obtained solution. Data collection and the refinement statistics are shown in Table I.
Negative-stain electron microscopy analysis
Purified WzzB (5 μL at 10 μg/mL) was applied to a glow-discharged carbon-coated EM grid for 1 min. Excess fluid was removed by blotting and the grid was washed for 15 s with a buffer containing Tris pH 7.5, 200 mM NaCl (no detergent). The sample was stained with 1.5% uranyl-formate (pH 5.0) for 1 min and the grid was dried. Digital electron micrographs were recorded with a FEI Tecnai F20 electron microscope (FEI) operated at 200 kV on a Gatan Ultrascan 4k × 4k Digital (CCD) System Camera (Gatan) at a calibrated magnification of 68,000×, corresponding to a pixel size of 2.2 Å at the specimen level and a defocus ranging from 1.5 to 2.5 µm. Single particles were manually selected using the program Boxer37 in 256 × 256 pixel boxes. The images were normalized then aligned using a reference free routine and an average was calculated. Processing was done using the SPARX software suite.38
Size-exclusion multiangle laser light scattering experiments
Size-exclusion chromatography with multiangle laser light-scattering (SEC-MALLS) experiments were performed using the Waters HPLC systems coupled to the Refractive Index and Static Laser Light-Scattering Instruments (Wyatt MiniDAWN Treos and Wyatt OptiLab rEX, respectively). The samples were first purified on Superdex-200 column in SEC buffer containing Tris pH 7.4, 200 mM NaCl, 5% (w/v) glycerol, 0.5 mM β-DDM, 1 mM TCEP to ensure their monodispersity and then injected onto the Superdex-200 column equilibrated with SEC buffer. A total of 50 μL (100 μg) of each protein was used for the SEC-MALS analysis injections. The data was processed using the Astra™ package (Wyatt Technologies).
Clear native PAGE and blue native PAGE analysis
Four to twelve percentage clear native gels were prepared using a gradient mixer. The light gel (4%) consisted of 375 mM Tris pH 8.8, 4% acrylamide, 0.05% APS, 0.05% β-DDM, while the heavy gel (12%) contained 375 mM Tris pH 8.8, 12% acrylamide, 0.05% APS (Ammonium Persulfate), 0.05% β-DDM. Tetramethylethylenediamine (TEMED) was added to 1/10th the volume of the APS just prior to poring the gels. The running buffer consisted of 250 mM Tris, and 1.92M glycine, pH ∼8.3. SDS and reducing agents were omitted from the 5× loading buffer, which contained 0.25M Tris pH 6.8, 50% (w/v) glycerol, and 0.25% (w/v) bromophenol blue. The gel was run for 3 h at 120 V at 4°C. For the blue native PAGE, the gel was prepared using the identical conditions except the running buffer contained 0.02% (w/v) Coomassie Blue G-250 dye. The gel was run for 45 min at 100V after which the running buffer was replaced by the clear-native PAGE running buffer and the gel was allowed to run for an additional 2 h at 180V. A total of 10 μg of each protein was loaded on each lane of the gel.
Acknowledgments
The research described in this paper was performed in part using CMCF beamline 08ID-1 at the Canadian Light Source, which is supported by the Natural Sciences and Engineering Research Council of Canada, the National Research Council Canada, the Canadian Institutes of Health Research, the Province of Saskatchewan, Western Economic Diversification Canada, and the University of Saskatchewan, and at the GM/CA-CAT beamline ID23 at the Advanced Photon Source, Argonne National Laboratory. The EM was performed at the Facility for Electron Microscopy Research at McGill University. They would like to thank Shaun Labiuk, Michel Fodje, Pawel Grochulski, Deqiang Yao, and Craig Ogata for copious personal help with crystal screening.
Glossary
- DDM
n-dodecyl-β-d-maltoside
- ECA
enterobacterial common antigen
- EM
electron microscope
- ET
Electron tomography
- LDAO
n-Dodecyl-N,N-Dimethylamine-N-Oxide
- LPS
lipopolysaccharide
- OG
octyl-β-d-glucopyranoside
- PCP
polysaccharide copolymerase
- PDB
Protein Data Bank
- SEC-MALLS
Size Exclusion Chromatography Multiangle Laser Light Scattering
- TM
transmembrane
Supporting Information
Additional Supporting Information may be found in the online version of this article.
Supporting Information
References
- 1.Whitfield C. Biosynthesis and assembly of capsular polysaccharides in Escherichia coli. Annu Rev Biochem. 2006;75:39–68. doi: 10.1146/annurev.biochem.75.103004.142545. [DOI] [PubMed] [Google Scholar]
- 2.Kalynych S, Morona R, Cygler M. Progress in understanding the assembly process of bacterial O-antigen. FEMS Microbiol Rev. 2014;38:1048–1065. doi: 10.1111/1574-6976.12070. [DOI] [PubMed] [Google Scholar]
- 3.Valvano MA. Export of O-specific lipopolysaccharide. Front Biosci. 2003;8:s452–s471. doi: 10.2741/1079. [DOI] [PubMed] [Google Scholar]
- 4.Smith AN, Boulnois GJ, Roberts IS. Molecular analysis of the Escherichia coli K5 kps locus: identification and characterization of an inner-membrane capsular polysaccharide transport system. Mol Microbiol. 1990;4:1863–1869. doi: 10.1111/j.1365-2958.1990.tb02035.x. [DOI] [PubMed] [Google Scholar]
- 5.Bronner D, Clarke BR, Whitfield C. Identification of an ATP-binding cassette transport system required for translocation of lipopolysaccharide O-antigen side-chains across the cytoplasmic membrane of Klebsiella pneumoniae serotype O1. Mol Microbiol. 1994;14:505–519. doi: 10.1111/j.1365-2958.1994.tb02185.x. [DOI] [PubMed] [Google Scholar]
- 6.Cuthbertson L, Kos V, Whitfield C. ABC transporters involved in export of cell surface glycoconjugates. Microbiol Mol Biol Rev. 2010;74:341–362. doi: 10.1128/MMBR.00009-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Greenfield LK, Whitfield C. Synthesis of lipopolysaccharide O-antigens by ABC transporter-dependent pathways. Carbohydr Res. 2012;356:12–24. doi: 10.1016/j.carres.2012.02.027. [DOI] [PubMed] [Google Scholar]
- 8.Morona R, Purins L, Tocilj A, Matte A, Cygler M. Sequence-structure relationships in polysaccharide co-polymerase (PCP) proteins. Trends Biochem Sci. 2009;34:78–84. doi: 10.1016/j.tibs.2008.11.001. [DOI] [PubMed] [Google Scholar]
- 9.Becker A, Puhler A. Specific amino acid substitutions in the proline-rich motif of the Rhizobium meliloti ExoP protein result in enhanced production of low-molecular-weight succinoglycan at the expense of high-molecular-weight succinoglycan. J Bacteriol. 1998;180:395–399. doi: 10.1128/jb.180.2.395-399.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Batchelor RA, Haraguchi GE, Hull RA, Hull SI. Regulation by a novel protein of the bimodal distribution of lipopolysaccharide in the outer membrane of Escherichia coli. J Bacteriol. 1991;173:5699–5704. doi: 10.1128/jb.173.18.5699-5704.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Barr K, Klena J, Rick PD. The modality of enterobacterial common antigen polysaccharide chain lengths is regulated by o349 of thewec gene cluster of Escherichia coliK-12. J Bacteriol. 1999;181:6564–6568. doi: 10.1128/jb.181.20.6564-6568.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wugeditsch T, Paiment A, Hocking J, Drummelsmith J, Forrester C, Whitfield C. Phosphorylation of Wzc, a tyrosine autokinase, is essential for assembly of group 1 capsular polysaccharides in Escherichia coli. J Biol Chem. 2001;276:2361–2371. doi: 10.1074/jbc.M009092200. [DOI] [PubMed] [Google Scholar]
- 13.Morona JK, Paton JC, Miller DC, Morona R. Tyrosine phosphorylation of CpsD negatively regulates capsular polysaccharide biosynthesis in streptococcus pneumoniae. Mol Microbiol. 2000;35:1431–1442. doi: 10.1046/j.1365-2958.2000.01808.x. [DOI] [PubMed] [Google Scholar]
- 14.Phoenix DA, Brandenburg K, Harris F, Seydel U, Hammerton T, Roberts IS. An investigation into the membrane-interactive potential of the Escherichia coli KpsE C-terminus. Biochem Biophys Res Commun. 2001;285:976–980. doi: 10.1006/bbrc.2001.5266. [DOI] [PubMed] [Google Scholar]
- 15.Larue K, Ford RC, Willis LM, Whitfield C. Functional and structural characterization of polysaccharide co-polymerase proteins required for polymer export in ATP-binding cassette transporter-dependent capsule biosynthesis pathways. J Biol Chem. 2011;286:16658–16668. doi: 10.1074/jbc.M111.228221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Papadopoulos M, Morona R. Mutagenesis and chemical cross-linking suggest that Wzz dimer stability and oligomerization affect lipopolysaccharide O-antigen modal chain length control. J Bacteriol. 2010;192:3385–3393. doi: 10.1128/JB.01134-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kintz EN, Goldberg JB. Site-directed mutagenesis reveals key residue for O antigen chain length regulation and protein stability in Pseudomonas aeruginosa Wzz2. J Biol Chem. 2011;286:44277–44284. doi: 10.1074/jbc.M111.273979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tran ENH, Morona R. Residues located inside the Escherichia coli FepE protein oligomer are essential for lipopolysaccharide O-antigen modal chain length regulation. Microbiology. 2013;159:701–714. doi: 10.1099/mic.0.065631-0. [DOI] [PubMed] [Google Scholar]
- 19.Daniels C, Morona R. Analysis of Shigella flexneri Wzz (Rol) function by mutagenesis and cross-linking: Wzz is able to oligomerize. Mol Microbiol. 1999;34:181–194. doi: 10.1046/j.1365-2958.1999.01591.x. [DOI] [PubMed] [Google Scholar]
- 20.Tang KH, Guo H, Yi W, Tsai MD, Wang PG. Investigation of the conformational states of Wzz and the Wzz.O-antigen complex under near-physiological conditions. Biochemistry. 2007;46:11744–11752. doi: 10.1021/bi701181r. [DOI] [PubMed] [Google Scholar]
- 21.Tocilj A, Munger C, Proteau A, Morona R, Purins L, Ajamian E, Wagner J, Papadopoulos M, Van Den BL, Rubinstein JL, Fethiere J, Matte A, Cygler M. Bacterial polysaccharide co-polymerases share a common framework for control of polymer length. Nat Struct Mol Biol. 2008;15:130–138. doi: 10.1038/nsmb.1374. [DOI] [PubMed] [Google Scholar]
- 22.Kalynych S, Yao D, Magee J, Cygler M. Structural characterization of closely related O-antigen lipopolysaccharide (LPS) chain length regulators. J Biol Chem. 2012;287:15696–15705. doi: 10.1074/jbc.M112.354837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Larue K, Kimber MS, Ford R, Whitfield C. Biochemical and structural analysis of bacterial O-antigen chain length regulator proteins reveals a conserved quaternary structure. J Biol Chem. 2009;284:7395–7403. doi: 10.1074/jbc.M809068200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stenberg F, Chovanec P, Maslen SL, Robinson CV, Ilag LL, von Heijne G, Daley DO. Protein complexes of the Escherichia coli cell envelope. J Biol Chem. 2005;280:34409–34419. doi: 10.1074/jbc.M506479200. [DOI] [PubMed] [Google Scholar]
- 25.Lasserre JP, Beyne E, Pyndiah S, Lapaillerie D, Claverol S, Bonneu M. A complexomic study of Escherichia coli using two-dimensional blue native/SDS polyacrylamide gel electrophoresis. Electrophoresis. 2006;27:3306–3321. doi: 10.1002/elps.200500912. [DOI] [PubMed] [Google Scholar]
- 26.Tong L, Rossmann MG. Rotation function calculations with GLRF program. In: Carter Jr., editor; Charles W, editor. Methods enzymol. New York: Academic Press; 1997. pp. 594–611. [PubMed] [Google Scholar]
- 27.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 28.Boekema EJ. Negative staining of integral membrane proteins. Micron Microscop Acta. 1991;22:319–462. [Google Scholar]
- 29.Marcia M, Ermler U, Peng G, Michel H. The structure of Aquifex aeolicus sulfide: quinone oxidoreductase, a basis to understand sulfide detoxification and respiration. Proc Natl Acad Sci USA. 2009;106:9625–9630. doi: 10.1073/pnas.0904165106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cherney MM, Zhang Y, Solomonson M, Weiner JH, James MN. Crystal structure of sulfide: quinone oxidoreductase from Acidithiobacillus ferrooxidans: insights into sulfidotrophic respiration and detoxification. J Mol Biol. 2010;398:292–305. doi: 10.1016/j.jmb.2010.03.018. [DOI] [PubMed] [Google Scholar]
- 31.Daniels C, Griffiths C, Cowles B, Lam JS. Pseudomonas aeruginosa O-antigen chain length is determined before ligation to lipid A core. Environ Microbiol. 2002;4:883–897. doi: 10.1046/j.1462-2920.2002.00288.x. [DOI] [PubMed] [Google Scholar]
- 32.Guo H, Lokko K, Zhang Y, Yi W, Wu Z, Wang PG. Overexpression and characterization of Wzz of Escherichia coli O86:H2. Protein Expr Purif. 2006;48:49–55. doi: 10.1016/j.pep.2006.01.015. [DOI] [PubMed] [Google Scholar]
- 33.Miroux B, Walker JE. Over-production of proteins in Escherichia coli: mutant hosts that allow synthesis of some membrane proteins and globular proteins at high levels. J Mol Biol. 1996;260:289–298. doi: 10.1006/jmbi.1996.0399. [DOI] [PubMed] [Google Scholar]
- 34.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 35.Adams PD, Afonine PV, Bunkoczi G, Chen VB, Echols N, Headd JJ, Hung LW, Jain S, Kapral GJ, Grosse Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner RD, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. The Phenix software for automated determination of macromolecular structures. Methods. 2011;55:94–106. doi: 10.1016/j.ymeth.2011.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J Appl Cryst. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ludtke SJ, Baldwin PR, Chiu W. EMAN: semiautomated software for high-resolution single-particle reconstructions. J Struct Biol. 1999;128:82–97. doi: 10.1006/jsbi.1999.4174. [DOI] [PubMed] [Google Scholar]
- 38.Hohn M, Tang G, Goodyear G, Baldwin PR, Huang Z, Penczek PA, Yang C, Glaeser RM, Adams PD, Ludtke SJ. SPARX, a new environment for Cryo-EM image processing. J Struct Biol. 2007;157:47–55. doi: 10.1016/j.jsb.2006.07.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information