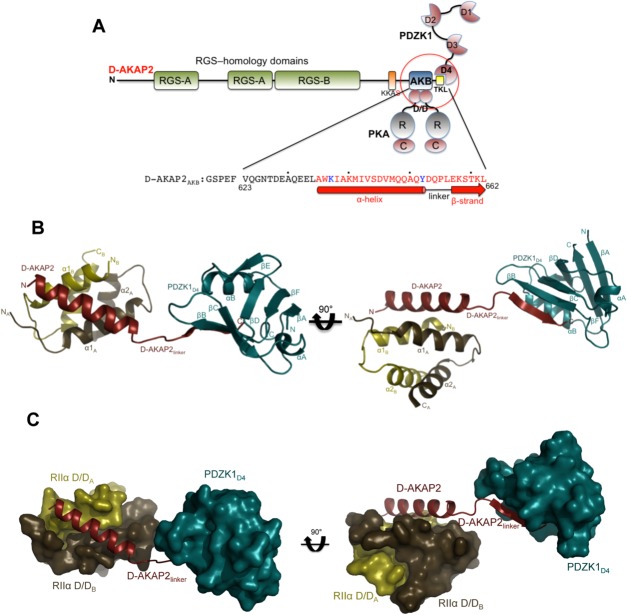
Figure 2.
Crystal structure of the D-AKAP2AKB: RIIα D/D: PDZK1D4 ternary complex. (a) Domain organization of D-AKAP2 and its interacting proteins. The RGS domains (RGS-A and RGS-B) interact with the small GTPases Rab4 and Rab11, the D-AKAP2AKB interacts with the D/D domains of PKA RI and RII subunits and the C-terminal type I PDZ motif, represented as –TKL, is known to interact with PDZK1 and NHERF1. The putative PKA phosphorylation site is shown as KKAS. The domains of the three proteins (D-AKAP2AKB, RIIα D/D, and PDZK1D4) used in this study are circled. The sequence of D-AKAP2AKB used for crystallization is shown and numbered, and every tenth residue indicated by a dot. Residues not modeled and modeled as an Ala in the structure are indicated in black and blue, respectively. Residues involved in an α-helix (interacting with RIIα D/D) and β-strand (interacting with PDZK1D4) are indicated by a solid bar and an arrow respectively. (b) Overall structure of the D-AKAP2AKB: RIIα D/D: PDZK1D4 ternary complex. The monomers of RIIα D/D are depicted in gold and brown, D-AKAP2AKB in red and PDZK1D4 in dark cyan. The N- and C-termini of each protein are indicated. D-AKAP2AKB has been divided into three clusters: the N-terminal region that interacts with RIIα D/D through an α-helix, the C-terminal region that interacts with PDZK1D4 through a β-strand and finally, the linker region that connects the N- and C-termini clusters. All structural figures were prepared using Pymol (http://www.pymol.org). (c) Surface representation of the ternary complex showing the importance of the D-AKAP2 linker in separating the RIIα D/D and PDZK1D4 molecules. For simplicity, the orientation and the coloring scheme match that of Figure 2b.