Fig 1.
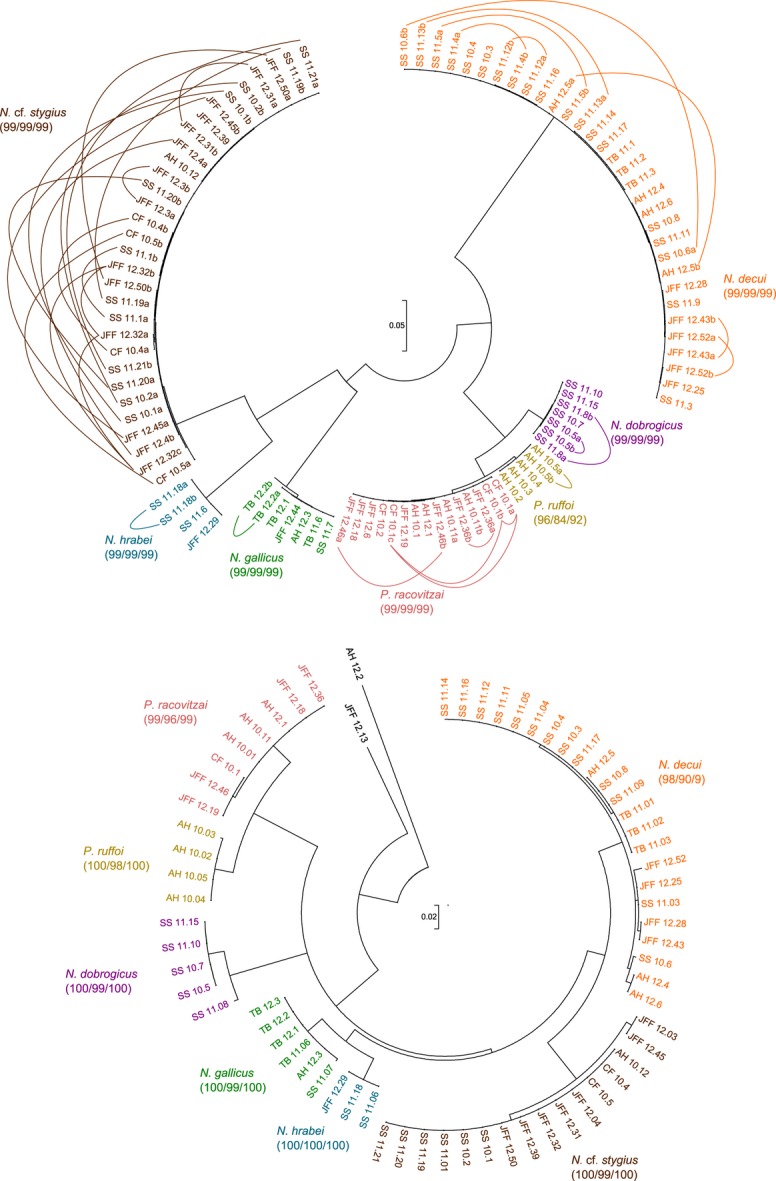
Top: Haploweb of ITS sequences of the 68 niphargid samples successfully sequenced for this marker. The two outgroups AH_12.2 (Synurella sp.) and JFF_12.13 (Gammarus sp.) were not included in the alignment, as their sequences were too divergent. The underlying phylogeny was obtained using a maximum-likelihood approach (model: GTR+G+I), following which connections were added between the sequences found co-occurring in heterozygous individuals. Bottom: Haplotree of COI sequences of the 67 amphipods samples successfully sequenced for this marker. The underlying phylogeny was obtained using a maximum-likelihood approach (model: GTR+G+I). Both approaches delineated seven species (represented by different colours); bootstrap values obtained from maximum-likelihood/parsimony/neighbour-joining are shown next to the name of each species.
