Figure 3.
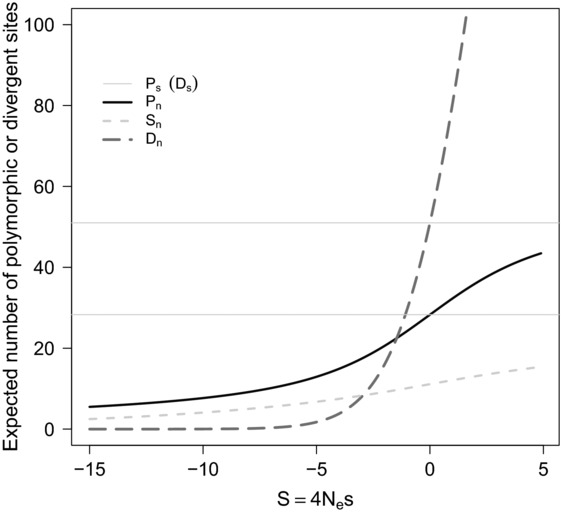
Expected amounts of polymorphism and divergence contributed by nonsynonymous mutations as a function of the scaled selection coefficient of a mutation. The expected amount of polymorphism in a genomic region can be summarized as the total number of polymorphic sites for both nonsynonymous (Pn) and synonymous sites (Ps, lower horizontal line), and the number of rare mutations using, for example, the number of singletons (mutations seen only once) at nonsynonymous sites (Sn). Amounts of divergence are quantified using the number of divergent synonymous (Ds, upper horizontal line) and nonsynonymous sites (Dn) relative to an out-group sequence. For a given mutation rate and effective population size, sample size n of chromosomes resequenced and size of a genomic fragment population genetic theory and diffusion results on the Wright-Fisher model can be used to compute these quantities as a function of the scaled mutation effect S of a nonsynonymous mutation. Here for illustration, we assume 1000 synonymous neutral sites, 3000 nonsynonymous nucleotide sites with a scaled mutation rate θ = 4Nμ = 0.01, a scaled divergence to the out-group of λ = 0.05, and a sample comprising n = 10 chromosomes.
