Abstract
Agrobacterium tumefaciens is a natural genetic engineer widely used to deliver DNA into various recipients, including plant, yeast and fungal cells. The bacterium can transfer single-stranded DNA molecules (T–DNAs) and bacterial virulence proteins, including VirE2. However, neither the DNA nor the protein molecules have ever been directly visualized after the delivery. In this report, we adopted a split-GFP approach: the small GFP fragment (GFP11) was inserted into VirE2 at a permissive site to create the VirE2-GFP11 fusion, which was expressed in A. tumefaciens; and the large fragment (GFP1–10) was expressed in recipient cells. Upon delivery of VirE2-GFP11 into the recipient cells, GFP fluorescence signals were visualized. VirE2-GFP11 was functional like VirE2; the GFP fusion movement could indicate the trafficking of Agrobacterium-delivered VirE2. As the natural host, all plant cells seen under a microscope received the VirE2 protein in a leaf-infiltration assay; most of VirE2 moved at a speed of 1.3–3.1 μm sec−1 in a nearly linear direction, suggesting an active trafficking process. Inside plant cells, VirE2-GFP formed filamentous structures of different lengths, even in the absence of T-DNA. As a non-natural host recipient, 51% of yeast cells received VirE2, which did not move inside yeast. All plant cells seen under a microscope transiently expressed the Agrobacterium-delivered transgene, but only 0.2% yeast cells expressed the transgene. This indicates that Agrobacterium is a more efficient vector for protein delivery than T-DNA transformation for a non-natural host recipient: VirE2 trafficking is a limiting factor for the genetic transformation of a non-natural host recipient. The split-GFP approach could enable the real-time visualization of VirE2 trafficking inside recipient cells.
Keywords: Agrobacterium, protein delivery, T-DNA, VirE2, visualization, Nicotiana benthamiana, Saccharomyces cerevisiae, technical advance
INTRODUCTION
As a natural genetic engineer, Agrobacterium tumefaciens can deliver T-DNA into different eukaryotes, including plant (Chilton et al., 1977; Marton et al., 1979; Broothaerts et al., 2005), yeast (Bundock et al., 1995; Piers et al., 1996), algae (Kathiresan et al., 2009) and fungal cells (de Groot et al., 1998). This is an efficient natural inter-kingdom transfer of genetic information. The use of A. tumefaciens as a genetic vector for different cells is becoming wide and diverse.
During the transfer process, a single-stranded DNA (T-DNA) molecule is generated inside the bacteria by VirD1-VirD2 endonuclease (Wang et al., 1984; Yanofsky et al., 1986; Scheiffele et al., 1995); afterwards, VirD2 remains covalently associated with the 5′ end of the T-DNA (T-strand; Yanofsky et al., 1986). This nucleoprotein complex is then transferred into recipient cells via a VirB/VirD4 type-IV secretion system (T4SS) (Cascales and Christie, 2004) in a manner mechanistically similar to a conjugation process (Beijersbergen et al., 1992).
The importance of T4SS is highlighted by the expanding list of bacterial pathogens that use T4SS to deliver proteins and nucleoprotein complexes into eukaryotic host cells (Cascales and Christie, 2003; Chen et al., 2004). An archetypal T4SS is the A. tumefaciens VirB/D4 apparatus (Cascales and Christie, 2004), which is responsible for T-DNA transfer from A. tumefaciens to natural host plants, as well as non-natural host recipients like bacteria, algae, yeast and fungal cells (Beijersbergen et al., 1992; Kelly and Kado, 2002; Cascales and Christie, 2003; Lacroix et al., 2006), in the presence of plant wound signal compounds, such as acetosyringone (AS; Stachel et al., 1985).
This T4SS is also known to translocate protein substrates, including a number of Vir proteins like VirE2, VirD2, VirE3, VirD5 and VirF (Vergunst et al., 2000, 2005; Schrammeijer et al., 2003). These translocated proteins are virulence effectors that interact with host factors in the recipient cells to facilitate transformation. As an abundant protein secreted into recipient cells, VirE2 is likely to coat and protect the T-strand (Citovsky et al., 1988, 1992; Yusibov et al., 1994; Rossi et al., 1996). VirE2 may also mediate the uptake of the T-DNA complex by forming a pore in the plant plasma membrane (Dumas et al., 2001). VirE2 could interact with plant VIP1, which is localized in the nucleus upon phosphorylation, and several importin α-isoforms in the plant cells, suggesting that VirE2 might also help with the nuclear targeting of T-DNA (Citovsky et al., 1992; Djamei et al., 2007; Bhattacharjee et al., 2008). In addition, VirE2 was reported to facilitate the chromatin targeting of the T-complex through an association with host VIP2 (Anand et al., 2007). It appears that VirE2 is involved in the entire T-complex trafficking inside recipient cells, from the entry point to the final destination.
Neither the DNA nor the protein molecules have ever been directly visualized inside the recipient cells after the translocation. It is not clear how the nucleoprotein complex is trafficked inside the recipient cells. In this report, we adopted a split-GFP system (Cabantous et al., 2005; Pedelacq et al., 2006) and successfully visualized VirE2, its aggregation forms and its movement in the recipient cells during a natural transformation process. When this was tested in different recipients, the data show that VirE2 delivery efficiency was comparable between a natural host and a non-natural host recipient, but the VirE2 trafficking and T-DNA transformation efficiency were very different. VirE2 moved in a nearly linear direction inside plant cells, whereas VirE2 did not move inside yeast. The T-DNA transformation efficiency for a natural host plant is 250–500-fold higher than for non-natural host yeast cells. This indicates that VirE2 trafficking inside recipient cells is a limiting factor for genetic transformation of a non-natural host recipient. Agrobacterium is more efficient in protein delivery than genetic transformation for a non-natural host recipient. The split-GFP approach enabled real-time visualization of VirE2 trafficking inside host cells.
RESULTS
A split-GFP approach to visualize VirE2
A split-GFP approach (Cabantous et al., 2005; Pedelacq et al., 2006; Van Engelenburg and Palmer, 2010) was used to visualize VirE2 inside recipient cells. As shown in Figure 1, the split-GFP system is composed of two non-fluorescent GFP fragments: β-strands 1–10 of GFP (GFP1–10), containing 215 amino acid residues, and β-strand 11 of GFP (GFP11), containing 16 amino acid residues. GFP1–10 and GFP11 could bind each other spontaneously and restore the fluorescence GFPcomp (Cabantous et al., 2005).
Figure 1.
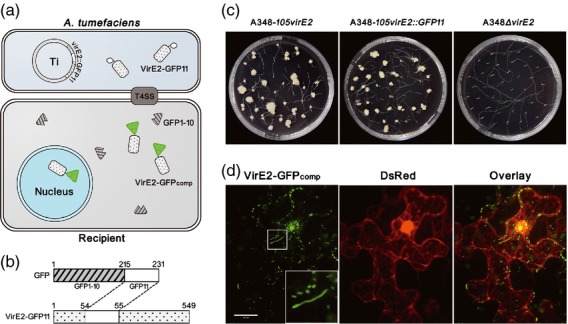
Visualization of VirE2 inside recipient cells using a split-GFP method. (a) General strategy for the visualization of Agrobacterium tumefaciens VirE2 delivered into recipient cells. VirE2-GFP11 encoded on Ti plasmid is expressed inside A. tumefaciens cells and is delivered into recipient cells, whereas GFP1–10 is expressed inside recipient cells. The GFP fluorescence complementation occurs upon VirE2-GFP11 translocation into recipient cells. (b) Construction of the VirE2-GFP11 fusion. The GFP11-coding sequence was inserted into virE2 at Pro54, and the fusion gene was then placed on the Ti plasmid by homologous recombination. (c) VirE2 virulence assay. The roots of transgenic Arabidopsis thaliana H16 expressing GFP1–10 were inoculated with A. tumefaciens cells with virE2 (A348-105virE2), virE2::GFP11 (A348-105virE2::GFP11) or virE2 deletion (A348ΔvirE2). Photographs were taken 4 weeks later. (d) Visualization of VirE2-GFPcomp inside plant cells. Cells of A. tumefaciens EHA105virE2::GFP11 were infiltrated into transgenic N. benthamiana (Nb308A) leaves expressing both GFP1–10 and DsRed. The leaf epidermal cells were observed at 2 days post-agroinfiltration under a confocal microscope with an Olympus UPLSAPO 60× NA 1.20 water immersion objective. DsRed expression facilitated the visualization of cellular locations. The boxed area is enlarged to highlight the structures of the VirE2-GFP11 complex. Scale bar: 20 μm.
We fused GFP11 onto VirE2 at a permissive site (Zhou and Christie, 1999) to create the VirE2-GFP11 fusion, the fusion was expressed inside A. tumefaciens, and GFP1–10 was expressed in the recipient cells. When VirE2-GFP11 was delivered into the recipient cells, GFP1–10 would be complemented by VirE2-GFP11 and the resulting VirE2-GFPcomp signals were detected.
As an abundant Vir protein (Engstrom et al., 1987), VirE2 is a non-specific single-stranded DNA (ssDNA) binding protein (Citovsky et al., 1988), which can coat the entire length of the T-strand in vitro with one VirE2 molecule, covering 19 bases of T-DNA (Citovsky et al., 1997). In the presence or absence of T-DNA, numerous VirE2 molecules can form telephone cord-like multimers in vitro (Citovsky et al., 1997; Frenkiel-Krispin et al., 2007; Dym et al., 2008). The self-association capacity could thus amplify the VirE2-GFPcomp signals to facilitate the direct visualization of VirE2 inside the recipient cells.
VirE2-GFP11 functions like VirE2
To ensure that the VirE2-GFP11 movement could represent VirE2 trafficking, the VirE2-GFP11 fusion should not disrupt the VirE2 function. To achieve this, GFP11 was inserted at Pro54 of VirE2 (accession no. AAZ50538), a site that was shown to be tolerant for a 31-residue oligopeptide insertion (Zhou and Christie, 1999). The virE2 gene from EHA105 was used to generate the VirE2-GFP fusion, as EHA105 does not contain any T-DNA (Hood et al., 1993), and thus VirE2 may be studied in the absence of T-DNA. To test the virulence function of VirE2-GFP, the fusion construct was then used to replace the virE2 gene of A348. The resulting A348-105virE2::GFP11 was inoculated onto roots of transgenic Arabidopsis thaliana (H16) expressing GFP1–10.
As shown in Figure 1c, A348-105virE2::GFP11 caused tumors just like the corresponding A348-105virE2, which is an A348 derivative with its virE2 replaced by EHA105 virE2. As expected, the virE2 deletion mutant A348ΔvirE2 was avirulent. The virulence function of VirE2-GFP11 was similar to the wild-type VirE2, as the frequency and size of tumors caused by A348-105virE2::GFP11 were similar to those of A348-virE2, even when different concentrations of the bacterial cells were used to test the virulence (Figure S1). In addition, the VirE2-GFP11 expressional level was similar to VirE2 (Figures S1 and S2); the GFP11 tagging did not affect bacterial growth (Figure S2). These results suggest that VirE2-GFP11 was fully functional, just like VirE2, even in the presence of GFP1–10 in the transgenic plants (Figure S1). Thus, the VirE2-GFP11 fusion was suitable for the visualization of VirE2 and its trafficking upon delivery into recipient cells.
Visualization of VirE2 movement inside Nicotiana benthamiana cells
To visualize VirE2 inside a natural host plant, A. tumefaciens EHA105virE2::GFP11 cells were infiltrated into transgenic N. benthamiana (Nb308A) leaves expressing both GFP1–10 and DsRed. When VirE2-GFP11 was translocated into the plant cells, GFP1–10 bound to VirE2-GFP11, and the resulting VirE2-GFPcomp signals appeared as green fluorescence under a confocal microscope (Figure 1d). The DsRed expression facilitated the visualization of the cellular locations. At 2 days after infiltration, VirE2-GFPcomp signals were found in the plant cells in both the cytoplasm and the nucleus (Figure 1d). Most of the signals appeared as spots, but some appeared as filamentous structures.
Time-course studies (Figure 2a) indicated that no VirE2-GFPcomp signals were detected at 16 h after agroinfiltration, presumably because it took time for the vir genes to be induced and for VirE2 to be delivered. In addition, a certain number of delivered VirE2-GFP11 molecules might be needed to form an aggregate that could subsequently become detectable, although the exact number is not clear. At 32 h after agroinfiltration, VirE2-GFPcomp signals were detected as spots. At 48 h after agroinfiltration, both VirE2-GFPcomp spots and filamentous structures were detected. Pre-induction of the bacteria by AS before agroinfiltration did not significantly speed up the appearance or increase the intensity of VirE2-GFPcomp signals. This suggests that vir gene induction was not a limiting factor, but it took time for VirE2 to be delivered to a detectable level in plant cells.
Figure 2.
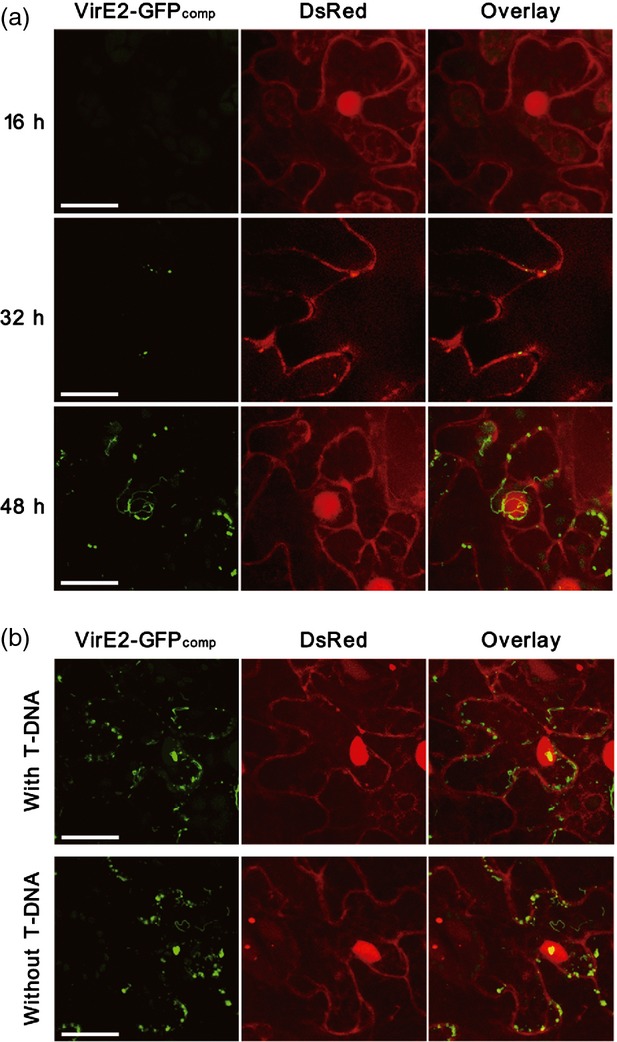
The time course of VirE2 delivery and the effect of T-DNA on VirE2 delivery. (a) The time course of VirE2 delivery in Nicotiana benthamiana. Agrobacterium tumefaciens EHA105virE2::GFP11 cells were infiltrated into transgenic N. benthamiana (Nb308A) leaves expressing both GFP1–10 and DsRed. The leaf epidermal cells were observed at 16, 32 and 48 h post-agroinfiltration. (b) The effect of T-DNA on VirE2 delivery. Cells of A. tumefaciens with T-DNA [EHA105virE2::GFP11(pHT101); upper panel] or without T-DNA (EHA105virE2::GFP11; lower panel) were infiltrated into transgenic N. benthamiana (Nb308A) leaves expressing both GFP1–10 and DsRed. The leaf epidermal cells were observed at 2 days post-agroinfiltration. Scale bars: 20 μm. Images were obtained under a confocal microscope with an Olympus UAPO N 340 40× NA 1.15 water immersion objective.
As the bacterial cells did not contain any T-DNA, the filamentous structures should be the aggregated form of the VirE2-GFP complex, free of any T-DNA. As the VirE2 transfer progressed, more filamentous structures were found and the filaments became even longer. This suggests that VirE2 aggregation grew when more VirE2 was delivered. Under the same conditions, the negative controls did not generate any GFP fluorescence; these included A. tumefaciens strains that did not encode VirE2-GFP11 or VirD4 (Figure S3). Therefore, naturally transferred VirE2 protein and its aggregated form were successfully visualized inside live recipient cells.
A binary plasmid pHT101 containing a T-DNA construct was introduced into the T-DNA-less EHA105virE2::GFP11 cells to study the effect of the T-strand on VirE2 delivery. As shown in Figure 2b, the presence or absence of the T-strand did not affect the quantity of VirE2 delivered or the formation of VirE2 filamentous structures inside the plant cells. This suggests that VirE2 delivery is independent of T-strand, which is consistent with previous observations (Binns et al., 1995; Sundberg et al., 1996).
VirE2 was reported to play a role in the T-complex trafficking by hijacking the plant MAPK-targeted VIP1 defense signaling pathway (Tzfira et al., 2002; Djamei et al., 2007). Thus we monitored the VirE2 movement in a time-lapse series, and successfully captured the VirE2-GFPcomp trafficking process (Movie S1). The speed of VirE2-GFP varied; the majority ranged from 1.3 to 3.1 μm sec−1 (Figure 3a). The movement was nearly linear and directional (Movie S1), suggesting that VirE2 movement was assisted by an active host process.
Figure 3.
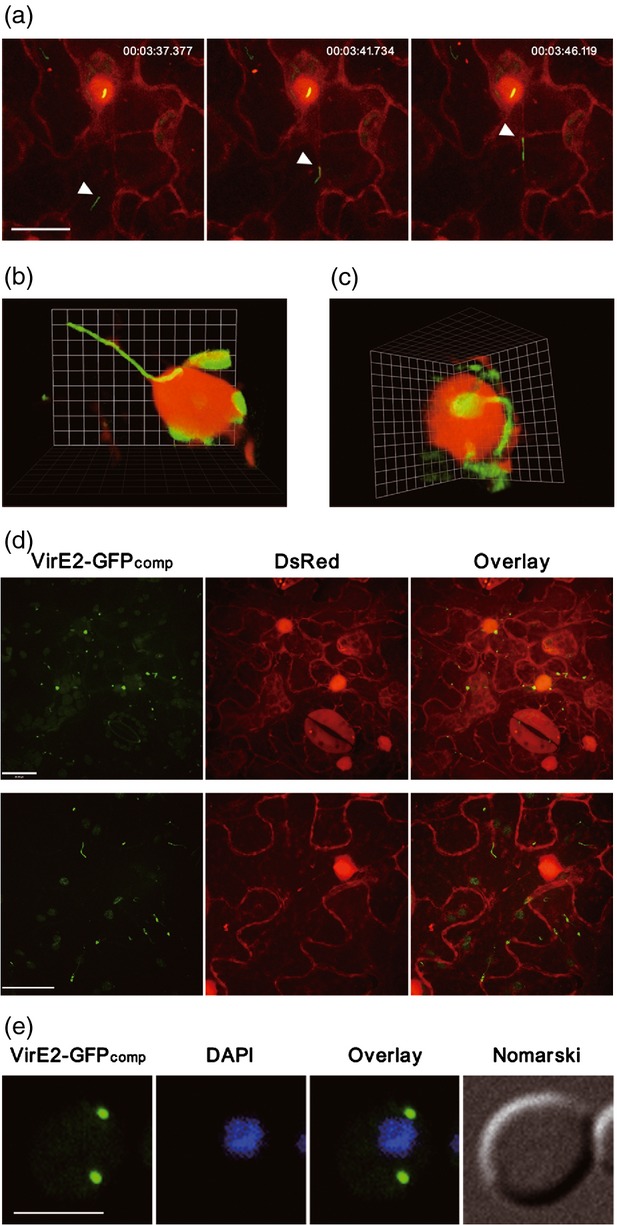
Tracking of VirE2 movement inside recipient cells. (a) VirE2-GFPcomp movement inside plant cells. Agrobacterium tumefaciens EHA105virE2::GFP11 cells were infiltrated into transgenic Nicotiana benthamiana (Nb308A) leaves expressing both GFP1–10 and DsRed. The movement of VirE2-GFPcomp signal (arrowed) inside the epidermal cells was monitored at 2 days post-agroinfiltration (Movie S1). Representative pictures from the time-lapse movie (Movie S1) show the VirE2 movement; a timer is shown at the top (h:min:sec). DsRed expression facilitated the visualization of cellular locations. Scale bar: 20 μm. (b) VirE2-GFPcomp complex attached onto the nucleus. (c) Filamentous VirE2-GFPcomp complex linked to VirE2-GFPcomp inside the nucleus. (d) Mutations at NLS1 of VirE2-GFP11 abolished the nuclear localization of VirE2-GFPcomp. Two representative fields (one in the upper panel; another in the lower panel) are chosen to show the effect of the mutations. Scale bar: 20 μm. (e) VirE2-GFPcomp signal inside yeast cells. Saccharomyces cerevisiae BY4741(pQH04-GFP1–10) cells were co-cultivated with AS-induced A. tumefaciens EHA105virE2::GFP11 and observed at 24 h post co-cultivation. Scale bar: 5 μm. All images were obtained under a confocal microscope with an Olympus UPLSAPO 60 × NA 1.20 water immersion objective.
Both the shorter and longer forms of VirE2 aggregation moved inside the plant cells; some movements were directed towards the nucleus. The VirE2-GFP filaments were found to be attached to the nucleus (Figure 3b; Movie S2). The filamentous VirE2-GFP complex was also found to be linked to the VirE2-GFP complex inside the nucleus (Figure 3c; Movie S3). These results suggest that the filamentous VirE2 complex was targeted for nuclear import. When the VirE2 nuclear localization signal 1 (NLS1; Citovsky et al., 1992) was mutated, the VirE2-GFP complex was exclusively localized in the cytoplasm and the nuclear import was not observed (Figure 3d) in either the VirE2-GFPcomp spots (upper panel) or the filamentous structures (lower panel). The VirE2-GFPcomp signals appeared to be less abundant when NLS1 was mutated, suggesting that the mutation might affect the stability or accumulation of the VirE2-GFPcomp complex. The experiments demonstrated that the nuclear import of the VirE2-GFP complex was dependent upon the nuclear localization signal.
VirE2 does not move inside yeast cells
Subsequently, this imaging approach was applied to a non-natural host species Saccharomyces cerevisiae. A yeast strain encoding GFP1–10 was co-cultivated with A. tumefaciens EHA105virE2::GFP11 that was induced by AS. VirE2-GFPcomp signals were visualized in yeast cells (Figure 3e), but not in the negative controls (Figure S4). The signals could appear as early as 2 h after co-cultivation (Figure S5a). However, the signals did not move inside the yeast cells (Figure S5b), and they were not localized in the nucleus (Figure 3e). More often they were at the periphery of the yeast cells. This indicates that VirE2 is not actively trafficked into yeast nuclei, presumably because yeast is a non-natural host recipient, and thus lacks the facilitator(s) for VirE2 trafficking. This is consistent with the previous observation that VirE2 was localized in the cytoplasm rather than in the nucleus of yeast cells (Rhee et al., 2000).
Comparison of VirE2 translocation and T-DNA transformation between N. benthamiana and yeast cells
We compared natural host plants with non-natural host yeast as recipients for Agrobacterium-mediated T-DNA transformation and VirE2 translocation. As shown in Figure 4, all N. benthamiana cells seen under the microscope received VirE2-GFPcomp signals, whereas 50.9% of S. cerevisiae cells received VirE2-GFPcomp signals. This indicates that the efficiency of Agrobacterium-mediated VirE2 translocation was comparable between natural host plant and non-natural host yeast. This suggests that VirE2 delivery into recipient cells is not a limiting factor for Agrobacterium-mediated transformation.
Figure 4.
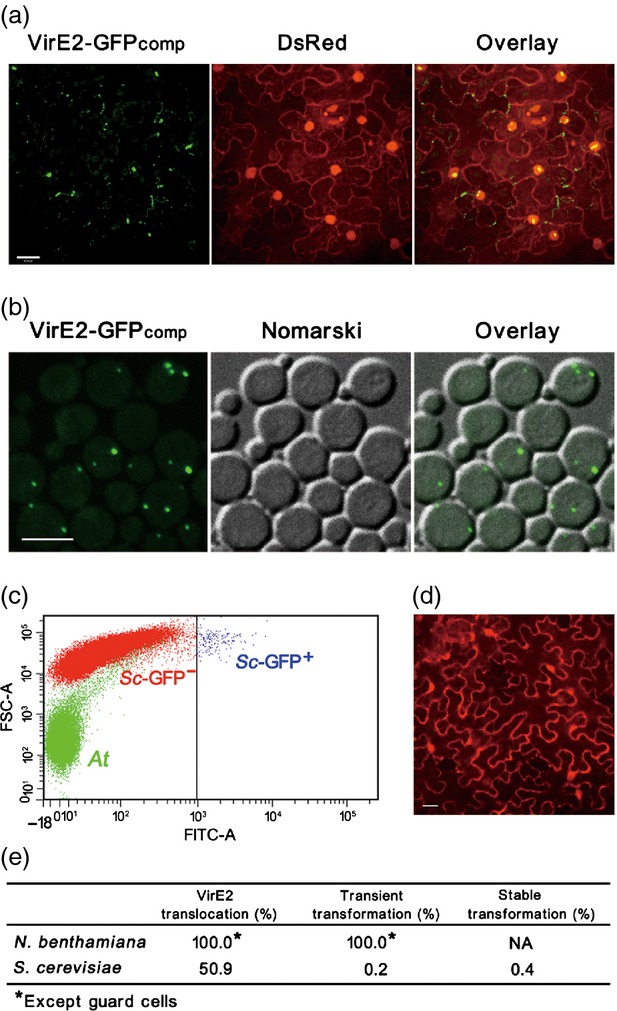
Comparison between plant and yeast as recipient cells for Agrobacterium-mediated transformation and VirE2 translocation. (a) Agrobacterium-mediated VirE2 translocation into plant cells. Agrobacterium tumefaciens EHA105virE2::GFP11 cells were infiltrated into transgenic Nicotiana benthamiana (Nb308A) leaves expressing both GFP1–10 and DsRed. The epidermal cells were observed at 2 days post-agroinfiltration under a confocal microscope with an Olympus UAPO N 340 40 × NA 1.15 water immersion objective. Scale bar: 20 μm. (b) Agrobacterium-mediated VirE2 translocation into yeast cells. Saccharomyces cerevisiae BY4741(pQH04-GFP1–10) cells were co-cultivated with AS-induced A. tumefaciens EHA105virE2::GFP11 and observed at 24 h post co-cultivation under a confocal microscope with an Olympus UPLSAPO 60 × NA 1.20 water immersion objective. Scale bar: 5 μm. (c) Determination of the efficiency of Agrobacterium-mediated T-DNA transfer into yeast cells. S. cerevisiae BY4741 cells were co-cultivated with AS-induced A. tumefaciens EHA105(pHT101) cells containing T-DNA harboring the EGFP reporter driven by the ADH promoter. At 24 h post co-cultivation, the cells were analyzed by FACS for the GFP intensity; At, A. tumefaciens cells; Sc-GFP−, untransformed S. cerevisiae cells; Sc-GFP+, S. cerevisiae cells expressing GFP. (d) Determination of the efficiency of Agrobacterium-mediated T-DNA transfer into plant cells. A. tumefaciens EHA105virE2::GFP11(pQH308A) cells were infiltrated into wild-type N. benthamiana leaves. The epidermal cells were observed at 2 days post agroinfiltration, under a confocal microscope with an Olympus UPlanSApo 20 × NA 0.60 water immersion objective. Scale bar: 20 μm. (e) Comparison between plant and yeast as recipient cells for Agrobacterium-mediated transformation and VirE2 translocation. The efficiency of Agrobacterium-mediated VirE2 translocation was determined based on the percentage of the cells showing VirE2-GFPcomp signal after agroinfiltration or co-cultivation. The efficiency of Agrobacterium-mediated transient transformation was determined based on the percentage of the cells showing DsRed or GFP signal from the T-DNA transferred into N. benthamiana or S. cerevisae. The efficiency of Agrobacterium-mediated stable transformation was determined based on the percentage of the cells carrying the T-DNA-encoded LEU2 transferred into S. cerevisae. NA, not applicable.
The efficiency of Agrobacterium-mediated transformation was dramatically different between natural host plants and the non-natural host yeast. As shown in Figure 4, all N. benthamiana cells seen under the microscope expressed the T-DNA-encoded DsRed in an infiltration assay for transient T-DNA transformation, whereas only 0.2% of S. cerevisiae cells expressed the T-DNA-encoded GFP in a co-cultivation assay by using a fluorescence-activated cell sorter (FACS) that can detect transient transformation in a timely manner. In an assay for stable transformation, 0.4% of S. cerevisiae cells were stably transformed with T-DNA that encodes a nutrient marker LEU2. The efficiency was similar between the transient and stable transformation for S. cerevisiae cells: the minor difference presumably arose from the sensitivity of the assay method.
The data indicate that the transient transformation efficiency of the natural host plant is 250–500-fold higher than the non-natural host yeast cells, whereas the VirE2 delivery was 127–255-fold more efficient than the transient transformation for a non-natural host recipient. One limiting factor for the non-natural host yeast transformation was presumably T-complex trafficking, as the non-natural host yeast cells could not facilitate the active trafficking of bacterial virulence factors, including VirE2 (Figure 3), and perhaps the T-strand.
DISCUSSION
Agrobacterium is widely used as a genetic vector to deliver DNA into various cells, whereas its capacity to deliver protein is not fully explored. In a sense Agrobacterium is regarded as a genetic engineer, and is not widely used as a vector for protein delivery. This article reports the direct visualization of Agrobacterium-delivered VirE2 protein inside recipient cells. The data indicate that Agrobacterium is more efficient in protein delivery than genetic transformation for a non-natural host recipient. It should be of significance to further explore the capacity of A. tumefaciens to deliver proteins.
The visualization of VirE2 inside recipient cells may also be useful to study the trafficking pathway of the T-strand, as VirE2 is a component of the proposed nucleoprotein complex. The abundance of VirE2 (Engstrom et al., 1987) is particularly suitable for its role to protect T-DNA by coating it with numerous molecules (Citovsky et al., 1988). It was estimated that about every 19 bases of T-DNA is coupled with one VirE2 molecule (Citovsky et al., 1997). In addition, the VirE2 protein can also assemble without T-DNA to form homodimers and solenoids (Frenkiel-Krispin et al., 2007). These unique traits prompted us to use VirE2 as a model to study Agrobacterium-delivered molecules inside recipient cells.
Previously, attempts have been made to tag VirE2 with different fluorescent proteins (Bhattacharjee et al., 2008; Aguilar et al., 2010). But none of the tagged VirE2 proteins was successfully translocated into recipient cells, presumably because the T4SS channel could not accommodate the enlarged size or hindering structure of these fusion proteins (Bhattacharjee et al., 2008; Aguilar et al., 2010). Nevertheless, the subcellular localization of VirE2 has been investigated with the tagged VirE2 protein; both cytoplasmic and nuclear localization of VirE2 have been reported (Citovsky et al., 1992; Rhee et al., 2000; Tzfira and Citovsky, 2001; Bhattacharjee et al., 2008). In those studies, VirE2 was artificially introduced into cells either by direct uptake or by transgenic expression, which may differ from a natural Agrobacterium-mediated transfer process. In addition, VirE2 tagged with a full-length GFP either at the C-terminus or N-terminus may affect its translocation activity (Simone et al., 2001; Christie et al., 2003; Schrammeijer et al., 2003; Bhattacharjee et al., 2008). This indicates that Agrobacterium-delivered VirE2 has never been visualized inside recipient cells previously.
In this report, we adopted a split GFP approach (Cabantous et al., 2005; Pedelacq et al., 2006), in which GFP is split into two parts: a small peptide (GFP11) of 16 residues and the remaining 215-residue fragment (GFP1–10). GFP11 was used as the tag and fused onto a permissive site of VirE2 so that the VirE2-GFP11 was functional like VirE2. This suggests that VirE2-GFP11 movement could represent the trafficking of VirE2. As GFP1–10 was expressed inside recipient cells, the GFPcomp signal was only detectable inside recipient cells, so that the VirE2-GFP11 trafficking signal could be readily monitored.
When GFP11 was fused onto the C-terminus of VirE2, no VirE2-GFPcomp signals were detected inside plant cells (Figure S6). This demonstrated that the position of GFP11 tagging was critical for the split-GFP experiments.
The split-GFP approach enabled us to directly visualize Agrobacterium-delivered VirE2 in live recipient cells. As VirE2-GFP11 is delivered in a natural setting, the VirE2-GFP11 movement should represent the natural trafficking process inside recipient cells. This should be of use to further study how host factors facilitate VirE2 movement inside recipient cells.
The nuclear import of VirE2 was reported to occur in plant cells (Citovsky et al., 1992, 1994; Tzfira and Citovsky, 2001), but not in yeast or mammalian cells (Guralnick et al., 1996; Rhee et al., 2000; Tzfira and Citovsky, 2001). Consistent with this, our split GFP assay demonstrated that VirE2-GFP moved towards the nucleus, and VirE2-GFP was indeed localized in the nucleus in the natural host leaves of N. benthamiana after agroinfiltration. Interestingly, we observed the nuclear targeting of the filamentous VirE2-GFP complex, which occurred in an NLS-dependent manner. The VirE2-GFP filaments grew in length as more VirE2-GFP was delivered, even in the absence of T-DNA. This is consistent with the fact that purified VirE2 can form filamentous structures in a self-aggregation manner (Frenkiel-Krispin et al., 2007; Dym et al., 2008).
The speed of VirE2-GFP movement varied; the majority ranged from 1.3 to 3.1 μm sec−1; the movement was linear and directional. This suggests that VirE2 trafficking inside plant cells is an active process. Currently, we are investigating the molecular mechanism of the VirE2 movement inside plant cells. Real-time visualization of VirE2 movement, as exemplified by the split-GFP approach, should be particularly useful to further dissect the detailed molecular events of VirE2 trafficking.
In contrast, VirE2 did not move and remained cytoplasmic, once it was delivered into the non-natural host yeast cells. Presumably, non-natural host recipient cells do not have the host factors to facilitate VirE2 movement. In addition, the filamentous VirE2-GFP structures were not observed inside yeast cells (Figure 3e). It is not clear whether this results from a limitation in yeast cellular space or from the quantity of VirE2 delivered into yeast cells.
Surprisingly, the efficiency of Agrobacterium-mediated VirE2 translocation into yeast is particularly high (50.9%) in our split-GFP experiments. In comparison, only 1% of S. cerevisiae cells received VirE2 in a Cre recombinase assay for translocation (CRAfT; Vergunst et al., 2000; Schrammeijer et al., 2003). The discrepancy may arise from the sensitivity of the detection methods. In our split-GFP assay, a small GFP fragment was inserted into a permissive site of VirE2; the structural disturbance from GFP tagging should be minimal. Indeed, the VirE2 fusion was functional like the wild type. On the other hand, the CRAfT assay used the Cre-VirE2 fusion, in which Cre has 343 residues, and this bulky size may reduce the efficiency of Cre-VirE2 delivery into the yeast cells. Moreover, our split-GFP imaging method can directly visualize VirE2 delivered into the cytoplasm, whereas the CRAfT assay might require a certain concentration of Cre-VirE2 delivered into the cells for Cre-mediated recombination to take place. Thus, the split-GFP imaging method should detect more signals than the CRAfT assay.
Our experiments showed that the efficiency of Agrobacterium-mediated VirE2 translocation was even similar between the natural host and non-natural host recipient. This indicates that Agrobacterium is a much more efficient vector for protein delivery than a genetic vector for a non-natural host recipient. The bacterial capability to deliver proteins should be further explored for both applied and theoretical investigations.
EXPERIMENTAL PROCEDURES
Strains, plasmids, primers and growth conditions
The strains and plasmids used in this study are listed in Table S1. A. tumefaciens strains were grown at 28°C in MG/L medium or induction medium (IBPO4; Cangelosi et al., 1991). S. cerevisiae strains were grown at 30°C using yeast extract peptone dextrose (YPD) medium (Clontech, now TaKaRa, http://www.takara-bio.com) or SD medium with appropriate drop-out (DO) supplements (Clontech). Escherichia coli strain DH5α was used for plasmid construction and was cultured at 37°C in lysogeny broth (LB) medium. Media were supplemented with 100 μg ml−1 carbenicillin or 100 μg ml−1 kanamycin, where needed.
Construction of plasmids and strains
Agrobacterium tumefaciens mutants were generated by adopting a sacB-based gene replacement strategy (Schweizer et al., 1998). A 1029-bp nptIII cassette from pCB301 (Oliver et al., 1999) was inserted into pEX18Tc (Schweizer et al., 1998) to generate Kmr-plasmid pEX18TcKm.
The GFP11-coding sequence was inserted into virE2 at Pro54 (of EHA105) using overlapping PCR. The resulting DNA fragment was inserted into pEX18TcKm to generate pEHA105-VE2::GFP11. This plasmid was then introduced into A. tumefaciens EHA105 by electroporation, and a sacB-based gene replacement was conducted. The resulting EHA105virE2::GFP11 was confirmed by PCR.
An EHA105 VirE2 nuclear localization signal 1 (NLS1) mutant was obtained by using the same strategy as described above. Eight residues of the NLS1 in EHA105 VirE2 (Citovsky et al., 1992), 221KLR…KYGRR237, were replaced with alanine. The resulting EHA105virE2::GFP11nls1 was confirmed by PCR.
To construct the A348 virE2 deletion mutant, a 834-bp fragment upstream of virE2 and a 985-bp fragment downstream of virE2 were amplified and inserted into pEX18TcKm to produce pA348-VE2KO, which was used to generate A348ΔvirE2.
The EHA105 virE2 or virE2::GFP11 coding sequence was inserted into pA348-VE2KO between the sequences upstream and downstream of A348 virE2 to generate pA348-105VE2 and pA348-105VE2::GFP11, respectively. These two plasmids were used to produce A348-105virE2 and A348-105virE2::GFP11.
To generate transgenic A. thaliana expressing GFP1–10, the coding sequence (American Peptide Company, http://www.americanpeptide.com) was inserted into pHB at HindIII (Mao et al., 2005) to produce pHB-GFP1–10.
To generate transgenic N. benthamiana expressing GFP1–10, the coding sequence (American Peptide Company) was inserted into pDs-Lox to replace Bar (Woody et al., 2007). The resulting cassette Pmas:GFP1–10:Tnos was amplified by PCR and inserted into pBI121 at ClaI-HindIII (Chen et al., 2003). A DsRed open reading frame (ORF) was then inserted at XbaI-BamHI, downstream of the 35S promoter, to generate pQH308A.
Generation of transgenic N. benthamiana and A. thaliana lines
The Agrobacterium-mediated transformation of N. benthamiana plants was performed using leaf sections. Transgenic calli were selected on MS media (Murashige and Skoog, 1962), supplemented with 100 mg l−1 kanamycin, 2 mg l−1 6-benzylaminopurine (6-BA), and 0.2 mg l−1 1-naphthaleneacetic acid. Transgenic tobacco plantlets were obtained by transferring calli with shoots into half-strength MS plates supplemented with 0.1 mg l−1 indole-3-butyric acid. The plasmid pQH308A was used to generate transgenic N. benthamiana Nb308A. Transgenic A. thaliana line H16 was generated with the floral-dip method (Clough and Bent, 1998).
Virulence assays
Arabidopsis thaliana seeds were surface-sterilized and then incubated at 4°C for 2 days. They were placed onto solidified half-strength MS medium supplemented with 1% sucrose and 0.5 g l−1 2-(N-morpholine)-ethanesulphonic acid (MES), pH 5.8. The plates were then incubated under a 16-h photoperiod at 25°C for 10–12 days. Roots from individual seedlings were cut into 3–5-mm segments and re-suspended in 1 ml of fresh half-strength MS medium containing A. tumefaciens cells at a concentration of 5 × 108 cell ml−1, except where specified otherwise. The mixtures were spread onto a solidified half-strength MS plate and subsequently incubated at 25°C for 2 days. The root segments were aligned onto half-strength MS medium plates containing 100 μg ml−1 cefotaxime and kept at 25°C for 4 weeks.
Agroinfiltration
To visualize Agrobacterium-delivered VirE2, agroinfiltration was performed as described previously (Lee and Yang, 2006). Briefly, the bacteria were grown overnight; the cultures were diluted 50 times in MG/L and then grown for 6 h. The bacteria were collected and re-suspended in infiltration buffer (10 mm MgCl2, 10 mm MES, pH 5.5) to OD600 = 1.0. The bacterial suspension was infiltrated using a syringe to the underside of fully expended N. benthamiana leaves. The infiltrated plant was maintained at 22°C with a photoperiod of 16 h of light/8 h of dark.
Agrobacterium-mediated transformation of yeast cells
The Agrobacterium-mediated transformation of yeast cells was performed as described by Piers et al. (1996), with modifications. Yeast cells were grown overnight; the cultures were diluted 40-fold with fresh YPD medium and grown for an additional 5 h. The cells were collected and the concentration was adjusted to 1 × 107 cells ml−1 in PBS. Agrobacteria were grown overnight; the cultures were diluted with fresh MG/L medium and grown for 7–8 h to reach the late log phase. The cells were collected and resuspended in induction medium with 100 μm AS. After growth at 28°C for 18 h, the cells were collected and resuspended in induction medium to a concentration of 1.2 × 1010 cells ml−1. Co-cultivation of bacteria and yeast cells was performed by mixing 50 μl of yeast cell suspension and 50 μl of A. tumefaciens cell suspension; the mixture was placed onto induction medium plates containing 100 μm AS and appropriate DO supplements. The cells were incubated at 20°C for 24 h before plating onto selection medium or before microscopic observation.
Confocal microscopy
A PerkinElmer UltraView Vox Spinning Disk system with electron-multiplying charge-coupled device (EM-CCD) cameras was used for confocal microscopy. To observe leaf epidermis, agroinfiltrated leaf tissues were detached from N. benthamiana plants and put in 2% low-melting agarose gel on a glass slide with a coverslip. For yeast images, the cells were suspended in PBS and then put on a slide with a coverslip. All images were taken in multiple focal planes (Z-stacks), and were processed to show the extended focus image or 3D opacity view by volocity® 3d image analysis software 6.2.1.
FACS analysis
Saccharomyces cerevisiae BY4741 cells were co-cultivated with AS-induced A. tumefaciens EHA105(pHT101) cells for 24 h. The cells were analyzed by FACSAria II (Becton-Dickinson, http://www.bd.com), based on the GFP intensity. A. tumefaciens EHA105 cells and S. cerevisiae BY4741 cells were used as negative controls.
Acknowledgments
The authors thank Dr Jian Xu for plasmid pHB and acknowledge the technical assistance from Yan Tong, Songci Xu, Lu Wee Tan and Yanbin Wang. This work is supported by grants from the Singapore Ministry of Education (R-154-000-437-112 and R-154-000-520-112).
SUPPORTING INFORMATION
Additional Supporting Information may be found in the online version of this article.
GFP11 tagging did not affect VirE2 function.
GFP11 tagging did not affect A. tumefaciens growth or VirE2 expression level.
Negative controls for the GFP fluorescence detected in N. benthamiana leaf epidermal cells under the experimental conditions for Figures 1–3.
Negative controls for the GFP fluorescence detected in yeast cells under the experimental conditions for Figures 2 and 3.
Visualization of VirE2-GFPcomp signals in yeast cells.
No detectable VirE2-GFPcomp signals for GFP11 fused onto the C-terminus of VirE2.
Strains and plasmids used in the studies.
VirE2 movement inside plant cells.
3D visualization of filamentous VirE2-GFPcomp attached onto the nucleus.
3D visualization of filamentous VirE2-GFPcomp complex linked to VirE2-GFPcomp inside the nucleus.
REFERENCES
- Aguilar J, Zupan J, Cameron TA, Zambryski PC. Agrobacterium type IV secretion system and its substrates form helical arrays around the circumference of virulence induced cells. Proc. Natl Acad. Sci. USA. 2010;107:3758–3763. doi: 10.1073/pnas.0914940107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anand A, Krichevsky A, Schomack S, Lahaye T, Tzfira T, Tang YH, Citovsky V, Mysore KS. Arabidopsis VirE2 Interacting Protein2 is required for Agrobacterium T-DNA integration in plants. Plant Cell. 2007;19:1695–1708. doi: 10.1105/tpc.106.042903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beijersbergen A, Dendulkras A, Schilperoort RA, Hooykaas PJJ. Conjugative transfer by the virulence system of Agrobacterium tumefaciens. Science. 1992;256:1324–1327. doi: 10.1126/science.256.5061.1324. [DOI] [PubMed] [Google Scholar]
- Bhattacharjee S, Lee LY, Oltmanns H, Cao H, Veena, Cuperus J, Gelvin SB. IMPa-4, an Arabidopsis importin alpha isoform, is preferentially involved in Agrobacterium-mediated plant transformation. Plant Cell. 2008;20:2661–2680. doi: 10.1105/tpc.108.060467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binns AN, Beaupre CE, Dale EM. Inhibition of VirB-mediated transfer of diverse substrates from Agrobacterium tumefaciens by the IncQ plasmid RSF1010. J. Bacteriol. 1995;177:4890–4899. doi: 10.1128/jb.177.17.4890-4899.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broothaerts W, Mitchell HJ, Weir B, Kaines S, Smith LMA, Yang W, Mayer JE, Roa-Rodriguez C, Jefferson RA. Gene transfer to plants by diverse species of bacteria. Nature. 2005;433:629–633. doi: 10.1038/nature03309. [DOI] [PubMed] [Google Scholar]
- Bundock P, Dendulkras A, Beijersbergen A, Hooykaas PJJ. Transkingdom T-DNA Transfer from Agrobacterium tumefaciens to Saccharomyces cerevisiae. EMBO J. 1995;14:3206–3214. doi: 10.1002/j.1460-2075.1995.tb07323.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabantous S, Terwilliger TC, Waldo GS. Protein tagging and detection with engineered self-assembling fragments of green fluorescent protein. Nat. Biotechnol. 2005;23:102–107. doi: 10.1038/nbt1044. [DOI] [PubMed] [Google Scholar]
- Cangelosi GA, Best EA, Martinetti G, Nester EW. Genetic analysis of Agrobacterium. Meth. Enzymol. 1991;204:384–397. doi: 10.1016/0076-6879(91)04020-o. [DOI] [PubMed] [Google Scholar]
- Cascales E, Christie PJ. The versatile bacterial type IV secretion systems. Nat. Rev. Microbiol. 2003;1:137–149. doi: 10.1038/nrmicro753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cascales E, Christie PJ. Definition of a bacterial type IV secretion pathway for a DNA substrate. Science. 2004;304:1170–1173. doi: 10.1126/science.1095211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen PY, Wang CK, Soong SC, To KY. Complete sequence of the binary vector pBI121 and its application in cloning T-DNA insertion from transgenic plants. Mol. Breeding. 2003;11:287–293. [Google Scholar]
- Chen J, de Felipe KS, Clarke M, Lu H, Anderson OR, Segal G, Shuman HA. Legionella effectors that promote nonlytic release from protozoa. Science. 2004;303:1358–1361. doi: 10.1126/science.1094226. [DOI] [PubMed] [Google Scholar]
- Chilton MD, Drummond MH, Merio DJ, Sciaky D, Montoya AL, Gordon MP, Nester EW. Stable incorporation of plasmid DNA into higher plant cells: the molecular basis of crown gall tumorigenesis. Cell. 1977;11:263–271. doi: 10.1016/0092-8674(77)90043-5. [DOI] [PubMed] [Google Scholar]
- Christie PJ, Atmakuri K, Ding ZY. VirE2, a type IV secretion substrate, interacts with the VirD4 transfer protein at cell poles of Agrobacterium tumefaciens. Mol. Microbiol. 2003;49:1699–1713. doi: 10.1046/j.1365-2958.2003.03669.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Citovsky V, Devos G, Zambryski P. Single-stranded DNA binding protein encoded by the virE locus of Agrobacterium tumefaciens. Science. 1988;240:501–504. doi: 10.1126/science.240.4851.501. [DOI] [PubMed] [Google Scholar]
- Citovsky V, Zupan J, Warnick D, Zambryski P. Nuclear localization of Agrobacterium VirE2 protein in plant cells. Science. 1992;256:1802–1805. doi: 10.1126/science.1615325. [DOI] [PubMed] [Google Scholar]
- Citovsky V, Warnick D, Zambryski P. Nuclear import of Agrobacterium VirD2 and VirE2 proteins in maize and tobacco. Proc. Natl Acad. Sci. USA. 1994;91:3210–3214. doi: 10.1073/pnas.91.8.3210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Citovsky V, Guralnick B, Simon MN, Wall JS. The molecular structure of Agrobacterium VirE2-single stranded DNA complexes involved in nuclear import. J. Mol. Biol. 1997;271:718–727. doi: 10.1006/jmbi.1997.1230. [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- Djamei A, Pitzschke A, Nakagami H, Rajh I, Hirt H. Trojan horse strategy in Agrobacterium transformation: abusing MAPK defense signaling. Science. 2007;318:453–456. doi: 10.1126/science.1148110. [DOI] [PubMed] [Google Scholar]
- Dumas F, Duckely M, Pelczar P, Van Gelder P, Hohn B. An Agrobacterium VirE2 channel for transferred-DNA transport into plant cells. Proc. Natl Acad. Sci. USA. 2001;98:485–490. doi: 10.1073/pnas.011477898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dym O, Albeck S, Unger T, Jacobovitch J, Brainzburg A, Michael Y, Frenkiel-Krispin D, Wolf SG, Elbaum M. Crystal structure of the Agrobacterium virulence complex VirE1-VirE2 reveals a flexible protein that can accommodate different partners. Proc. Natl Acad. Sci. USA. 2008;105:11170–11175. doi: 10.1073/pnas.0801525105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engstrom P, Zambryski P, Van Montagu M, Stachel S. Characterization of Agrobacterium tumefaciens virulence proteins induced by the plant factor acetosyringone. J. Mol. Biol. 1987;197:635–645. doi: 10.1016/0022-2836(87)90470-0. [DOI] [PubMed] [Google Scholar]
- Frenkiel-Krispin D, Wolf SG, Albeck S, et al. Plant transformation by Agrobacterium tumefaciens – Modulation of single-stranded DNA-VirE2 complex assembly by VirE1. J. Biol. Chem. 2007;282:3458–3464. doi: 10.1074/jbc.M605270200. [DOI] [PubMed] [Google Scholar]
- de Groot MJA, Bundock P, Hooykaas PJJ, Beijersbergen AGM. Agrobacterium tumefaciens-mediated transformation of filamentous fungi. Nat. Biotechnol. 1998;16:839–842. doi: 10.1038/nbt0998-839. [DOI] [PubMed] [Google Scholar]
- Guralnick B, Thomsen G, Citovsky V. Transport of DNA into the nuclei of xenopus oocytes by a modified VirE2 protein of Agrobacterium. Plant Cell. 1996;8:363–373. doi: 10.1105/tpc.8.3.363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hood EE, Gelvin SB, Melchers LS, Hoekema A. New Agrobacterium helper plasmids for gene transfer to plants. Transgenic Res. 1993;2:208–218. [Google Scholar]
- Kathiresan S, Chandrashekar A, Ravishankar GA, Sarada R. Agrobacterium-mediated transformation in the green alga Haematococcus pluvialis (Chlorophyceae, Volvocales) J. Phycol. 2009;45:642–649. doi: 10.1111/j.1529-8817.2009.00688.x. [DOI] [PubMed] [Google Scholar]
- Kelly BA, Kado CI. Agrobacterium-mediated T-DNA transfer and integration into the chromosome of Streptomyces lividans. Mol. Plant Pathol. 2002;3:125–134. doi: 10.1046/j.1364-3703.2002.00104.x. [DOI] [PubMed] [Google Scholar]
- Lacroix B, Tzfira T, Vainstein A, Citovsky V. A case of promiscuity: Agrobacterium's endless hunt for new partners. Trends Genet. 2006;22:29–37. doi: 10.1016/j.tig.2005.10.004. [DOI] [PubMed] [Google Scholar]
- Lee MW, Yang Y. Transient expression assay by agroinfiltration of leaves. Methods Mol. Biol. 2006;323:225–229. doi: 10.1385/1-59745-003-0:225. [DOI] [PubMed] [Google Scholar]
- Mao J, Zhang YC, Sang Y, Li QH, Yang HQ. A role for Arabidopsis cryptochromes and COP1 in the regulation of stomatal opening. Proc. Natl Acad. Sci. USA. 2005;102:12270–12275. doi: 10.1073/pnas.0501011102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marton L, Wullems GJ, Molendijk L, Schilperoort RA. In vitro transformation of cultured cells from Nicotiana tabacum by Agrobacterium tumefaciens. Nature. 1979;277:129–131. [Google Scholar]
- Murashige T, Skoog F. A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol. Plant. 1962;15:473–497. [Google Scholar]
- Oliver DJ, Xiang CB, Han P, Lutziger I, Wang K. A mini binary vector series for plant transformation. Plant Mol. Biol. 1999;40:711–717. doi: 10.1023/a:1006201910593. [DOI] [PubMed] [Google Scholar]
- Pedelacq JD, Cabantous S, Tran T, Terwilliger TC, Waldo GS. Engineering and characterization of a superfolder green fluorescent protein. Nat. Biotechnol. 2006;24:79–88. doi: 10.1038/nbt1172. [DOI] [PubMed] [Google Scholar]
- Piers KL, Heath JD, Liang XY, Stephens KM, Nester EW. Agrobacterium tumefaciens-mediated transformation of yeast. Proc. Natl Acad. Sci. USA. 1996;93:1613–1618. doi: 10.1073/pnas.93.4.1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhee Y, Gurel F, Gafni Y, Dingwall C, Citovsky V. A genetic system for detection of protein nuclear import and export. Nat. Biotechnol. 2000;18:433–437. doi: 10.1038/74500. [DOI] [PubMed] [Google Scholar]
- Rossi L, Hohn B, Tinland B. Integration of complete transferred DNA units is dependent on the activity of virulence E2 protein of Agrobacterium tumefaciens. Proc. Natl Acad. Sci. USA. 1996;93:126–130. doi: 10.1073/pnas.93.1.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheiffele P, Pansegrau W, Lanka E. Initiation of Agrobacterium tumefaciens T-DNA processing: purified proteins VirD1 and VirD2 catalyze site-specific and strand-specific cleavage of superhelical T-border DNA in vitro. J. Biol. Chem. 1995;270:1269–1276. doi: 10.1074/jbc.270.3.1269. [DOI] [PubMed] [Google Scholar]
- Schrammeijer B, den Dulk-Ras A, Vergunst AC, Jacome EJ, Hooykaas PJJ. Analysis of Vir protein translocation from Agrobacterium tumefaciens using Saccharomyces cerevisiae as a model: evidence for transport of a novel effector protein VirE3. Nucleic Acids Res. 2003;31:860–868. doi: 10.1093/nar/gkg179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweizer HP, Hoang TT, Karkhoff-Schweizer RR, Kutchma AJ. A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally-located DNA sequences: application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene. 1998;212:77–86. doi: 10.1016/s0378-1119(98)00130-9. [DOI] [PubMed] [Google Scholar]
- Simone M, McCullen CA, Stahl LE, Binns AN. The carboxy-terminus of VirE2 from Agrobacterium tumefaciens is required for its transport to host cells by the virB-encoded type IV transport system. Mol. Microbiol. 2001;41:1283–1293. doi: 10.1046/j.1365-2958.2001.02582.x. [DOI] [PubMed] [Google Scholar]
- Stachel SE, Messens E, Vanmontagu M, Zambryski P. Identification of the signal molecules produced by wounded plant cells that activate T-DNA transfer in Agrobacterium tumefaciens. Nature. 1985;318:624–629. [Google Scholar]
- Sundberg C, Meek L, Carroll K, Das A, Ream W. VirE1 protein mediates export of the single-stranded DNA-binding protein VirE2 from Agrobacterium tumefaciens into plant. J. Bacteriol. 1996;178:1207–1212. doi: 10.1128/jb.178.4.1207-1212.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzfira T, Citovsky V. Comparison between nuclear localization of nopaline- and octopine-specific Agrobacterium VirE2 proteins in plant, yeast and mammalian cells. Mol. Plant Pathol. 2001;2:171–176. doi: 10.1046/j.1364-3703.2001.00065.x. [DOI] [PubMed] [Google Scholar]
- Tzfira T, Vaidya M, Citovsky V. Increasing plant susceptibility to Agrobacterium infection by overexpression of the Arabidopsis nuclear protein VIP1. Proc. Natl Acad. Sci. USA. 2002;99:10435–10440. doi: 10.1073/pnas.162304099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Engelenburg SB, Palmer AE. Imaging type-III secretion reveals dynamics and spatial segregation of Salmonella effectors. Nat. Methods. 2010;7:325–330. doi: 10.1038/nmeth.1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vergunst AC, Schrammeijer B, den Dulk-Ras A, de Vlaam CMT, Regensburg-Tuink TJG, Hooykaas PJJ. VirB/D4-dependent protein translocation from Agrobacterium into plant cells. Science. 2000;290:979–982. doi: 10.1126/science.290.5493.979. [DOI] [PubMed] [Google Scholar]
- Vergunst AC, van Lier MCM, den Dulk-Ras A, Stuve TAG, Ouwehand A, Hooykaas PJJ. Positive charge is an important feature of the C-terminal transport signal of the VirB/D4-translocated proteins of Agrobacterium. Proc. Natl Acad. Sci. USA. 2005;102:832–837. doi: 10.1073/pnas.0406241102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Herrera-Estrella L, Van Montagu M, Zambryski P. Right 25 bp terminus sequence of the nopaline T-DNA is essential for and determines direction of DNA transfer from Agrobacterium to the plant genome. Cell. 1984;38:455–462. doi: 10.1016/0092-8674(84)90500-2. [DOI] [PubMed] [Google Scholar]
- Woody ST, Austin-Phillips S, Amasino RM, Krysan PJ. The WiscDsLox T-DNA collection: an arabidopsis community resource generated by using an improved high-throughput T-DNA sequencing pipeline. J. Plant. Res. 2007;120:157–165. doi: 10.1007/s10265-006-0048-x. [DOI] [PubMed] [Google Scholar]
- Yanofsky MF, Porter SG, Young C, Albright LM, Gordon MP, Nester EW. The virD2 operon of Agrobacterium tumefaciens encodes a site-specific endonuclease. Cell. 1986;47:471–477. doi: 10.1016/0092-8674(86)90604-5. [DOI] [PubMed] [Google Scholar]
- Yusibov VM, Steck TR, Gupta V, Gelvin SB. Association of single-stranded transferred DNA from Agrobacterium tumefaciens with tobacco cells. Proc. Natl Acad. Sci. USA. 1994;91:2994–2998. doi: 10.1073/pnas.91.8.2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou XR, Christie PJ. Mutagenesis of the Agrobacterium VirE2 single-stranded DNA-binding protein identifies regions required for self-association and interaction with VirE1 and a permissive site for hybrid protein construction. J. Bacteriol. 1999;181:4342–4352. doi: 10.1128/jb.181.14.4342-4352.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
GFP11 tagging did not affect VirE2 function.
GFP11 tagging did not affect A. tumefaciens growth or VirE2 expression level.
Negative controls for the GFP fluorescence detected in N. benthamiana leaf epidermal cells under the experimental conditions for Figures 1–3.
Negative controls for the GFP fluorescence detected in yeast cells under the experimental conditions for Figures 2 and 3.
Visualization of VirE2-GFPcomp signals in yeast cells.
No detectable VirE2-GFPcomp signals for GFP11 fused onto the C-terminus of VirE2.
Strains and plasmids used in the studies.
VirE2 movement inside plant cells.
3D visualization of filamentous VirE2-GFPcomp attached onto the nucleus.
3D visualization of filamentous VirE2-GFPcomp complex linked to VirE2-GFPcomp inside the nucleus.


