Abstract
Light is essential for plant survival; as such, plants flexibly adjust their growth and development to best harvest light energy. Brassinosteroids (BRs), plant growth-promoting steroid hormones, are essential for this plasticity of development. However, the precise mechanisms underlying BR-mediated growth under different light conditions remain largely unknown. Here, we show that darkness increases the activity of the BR-specific transcription factor, BZR1, by decreasing the phosphorylated (inactive) form of BZR1 in a proteasome-dependent manner. We observed that COP1, a dark-activated ubiquitin ligase, captures and degrades the inactive form of BZR1. In support of this, BZR1 is abundant in the cop1-4 mutant. The removal of phosphorylated BZR1 in darkness increases the ratio of dephosphorylated to phosphorylated forms of BZR1, thus increasing the chance of active homodimers forming between dephosphorylated BZR1 proteins. Furthermore, a transcriptome analysis revealed the identity of genes that are likely to contribute to the differential growth of hypocotyls in light conditions. Transgenic misexpression of three genes under the 35S promoter in light conditions resulted in elongated petioles and hypocotyls. Our results suggest that light conditions directly control BR signaling by modulating BZR1 stability, and consequently by establishing light-dependent patterns of hypocotyl growth in Arabidopsis.
Keywords: brassinosteroids, photomorphogenesis, hypocotyl elongation, proteasome, brassinazole, Arabidopsis thaliana
INTRODUCTION
The ability of plants to adapt their growth patterns to different light conditions underscores an important survival strategy in natural environments. When not exposed to optimal light, plants mobilize resources to grow and reach the light as part of a developmental process termed the shade avoidance syndrome (SAS) (Morelli and Ruberti, 2002; Franklin and Whitelam, 2005; Vandenbussche et al., 2005). Arabidopsis seedlings exhibiting SAS have longer hypocotyls and smaller leaf blades than their light-grown counterparts. Growth-promoting plant hormones, including auxin and gibberellins, control SAS (Pierik et al., 2004; Alabadi et al., 2008; Feng et al., 2008; de Lucas et al., 2008; Tao et al., 2008; Stavang et al., 2009). Many genes are known to be involved in SAS (Carabelli et al., 1996; Steindler et al., 1999; Kozuka et al., 2005; Bou-Torrent et al., 2008; Sorin et al., 2009; Leivar and Quail, 2011). Arabidopsis loss-of-function phytochrome B (phyB) mutants (Bae and Choi, 2008) and PHYTOCHROME INTERACTING FACTOR (PIF) overexpression lines constitutively display SAS in the light (Kim et al., 2003), suggesting that these genes control the growth pattern under various light conditions.
Brassinosteroids (BRs) are growth-promoting plant steroid hormones, and thus mutants defective in BR biosynthesis or signaling display characteristic growth-deficiency phenotypes (Clouse et al., 1996; Li et al., 1996; Szekeres et al., 1996; Choe et al., 1998, 1999a,b, 2000). Interestingly, SAS phenotypes of phyB disappear when combined with mutations defective in BR biosynthesis (Luccioni et al., 2002), suggesting that BRs are implicated in phyB-mediated hypocotyl growth. However, the mechanism by which light conditions control the degree of BR activity remains elusive. A current model depends on the interplay of the two transcription factors: PIF4 and BRASSINAZOLE RESISTANT 1 (BZR1) (Oh et al., 2012). When seedlings elongate in darkness, PIF4 and BZR1 form a heterodimer that regulates the transcription of their target genes; GERMIN-LIKE PROTEIN 1 (GER1) is repressed and PACLOBUTRAZOL RESISTANCE genes (PRE5 and PRE6) are activated (Oh et al., 2012). As PIF4 is a light-labile protein (Lorrain et al., 2008), BZR1 stimulates growth more effectively in darkness. This model is also supported by the phenotype of the gain-of-function mutant, bzr1-D, which shows resistance to a BR biosynthetic inhibitor only in darkness (Wang et al., 2002). It is likely that the deficiency of PIF4 in light conditions limits the growth-promoting effect of bzr1-D. As such, BRs seem to participate in the differential growth of hypocotyls in part through their interaction with PIF4. However, it is not known how the activity of BZR1 itself is differentially controlled in light and darkness.
BZR1 is a substrate of a glycogen synthase kinase 3β-like kinase, BR INSENSITIVE 2 (BIN2; Choe et al., 2002; Li and Nam, 2002; Li et al., 2001; Perez-Perez et al., 2002). Upon phosphorylation, BZR1 is captured by 14-3-3 proteins and sequestered in the cytoplasm (Gampala et al., 2007; Ryu et al., 2007), or its DNA binding affinity is reduced as part of an inactivation mechanism (Vert and Chory, 2006). Thus, the ratio of dephosphorylated to phosphorylated BZR1 serves as an index to estimate the signaling status of the plant (Wang et al., 2002). The phosphorylation status of BZR1 does not seem to change under different light conditions (He et al., 2002; Wang et al., 2002), however, and the hypocotyls of the quadruple mutant of pif1-4 (pifq) still elongate in response to darkness (Shin et al., 2009; Leivar and Quail, 2011), suggesting that a mechanism that directly regulates BZR1 activity exists and remains to be identified.
During our quest to discover the mechanisms that govern BZR1 activity, we found that the gulliver2/phyB-77 mutant possesses a reduced level of phosphorylated BZR1 in light. Surprisingly, we found that when plants are transferred from light to darkness, the protein levels of BZR1, mostly the phosphorylated forms, are gradually degraded in a proteasome-dependent manner. In vivo pull-down analyses revealed that CONSTITUTIVE PHOTOMORPHOGENIC 1 (COP1), an E3 ligase, binds to phosphorylated forms of BZR1. In addition, transcriptome analyses of phyB-77, bri1-5 and the phyB-77 bri1-5 double mutant revealed the unique set of genes that is likely to be regulated by BZR1 under the direction of both BR and light signaling. We show that light and darkness directly control components of the BR signaling pathway to confer optimal growth patterns according to the specific light conditions.
RESULTS AND DISCUSSION
Upregulation of feedback downregulation signaling in the phytochrome B-77 mutant
To understand the molecular mechanisms underlying the interaction between BRs and light, we isolated gulliver 2 (gul2) mutants by screening Arabidopsis mutant populations for elongated hypocotyls in the presence of both a BR biosynthetic inhibitor, brassinazole (Brz), and light. We isolated two alleles: gul2-1 and gul2-2. The gul2-1 mutant was in the Wassilewskija-2 (Ws-2) background and was mutagenized by ethyl methanesulfonate (EMS), whereas gul2-2 was obtained from a T-DNA activation-tagged mutant population in the Columbia-0 (Col-0) ecotype (Kim et al., 2014). Through the combined methods of map-based cloning and candidate gene sequencing, we revealed that both gul2-1 and gul2-2 mutants have defects in PHYTOCHROME B (PhyB), so they were renamed as phyB-77 and phyB-78, respectively. The phyB-77/gul2-1 mutant had a single base-pair substitution that changed Gln at the 438th residue to a premature stop codon. The untagged phyB-78/gul2-2 allele had a deletion mutation at 582–613 bp from the start codon, which caused a frameshift mutation, eventually introducing a premature stop codon at amino acid 196.
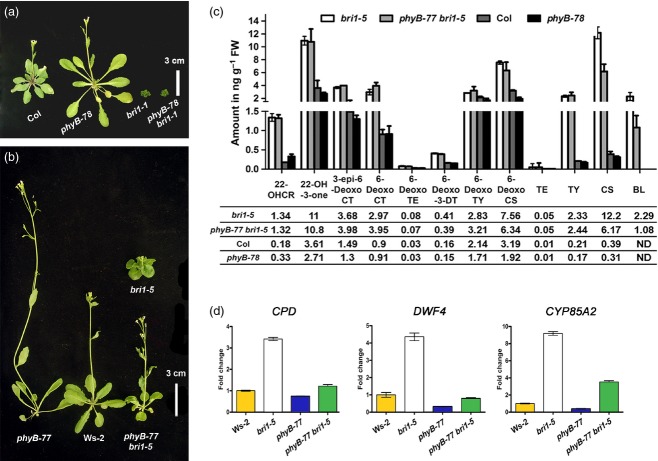
To learn how BR signaling interacts with this gul2/phyB, we first examined the epistatic interaction between phyB and a weak brassinosteroid insensitive 1 mutant bri1-5 (Noguchi et al., 1999). The phenotype of a double mutant of the strong allele bri1-1 and phyB-78 was indistinguishable from that of the bri1-1 single mutant, whereas the weak allele bri1-5 was dramatically suppressed by phyB-77 mutation (Figure 1a,b), suggesting that BR signaling is essential for light-dependent growth. The elongated phenotype of the phyB-77 single or phyB-77 bri1-5 double mutant could stem from increased BR biosynthesis or signaling, or both. Therefore, we quantified endogenous BRs and noticed that the levels of the most bioactive BR, brassinolide (BL), were significantly lower in the phyB-77 bri1-5 double mutant than in the bri1-5 single mutant (Figure 1c). As a reduced level of BL is considered a consequence of increased BR signaling activity, mediated by BZR1 (Choe et al., 2001; Wang et al., 2002; Chung et al., 2011, 2012; Kim et al., 2013), the reduction in bioactive BR levels in phyB-78 and phyB-77 bri1-5 mutants implies that feedback downregulation signaling is activated by phyB. Consistently, the transcript levels of BR biosynthetic genes were low in phyB-77 bri1-5 mutant seedlings (Figure 1d). The transcript levels of CONSTITUTIVE PHOTOMORPHOGENESIS AND DWARFISM (CPD), DWARF 4 (DWF4) and CYP85A2 were high in bri1-5, but were suppressed in the double mutant, suggesting that feedback downregulation mechanisms are functional in the double mutant because of phyB-77.
Figure 1.
Activation of brassinosteroid (BR) signaling in a weak bri1-5 allele by the phyB mutation. (a) A strong bri1-1 allele is epistatic to phyB-78. (b) A weak bri1-5 allele is suppressed by phyB-77. The dwarfism of bri1-5 was suppressed by phyB-77; the double mutant and the wild type were similar in stature. The plants were grown for 6 weeks under long-day conditions (16-h light/8-h dark), with a photon fluence rate of 80 μmol m−2 s−1 at 22°C. (c) Differences in BR levels in 5-week-old wild type, bri1-5, phyB-77 and phyB-77 bri1-5 plants. Tissues were collected from aerial parts. Mean values of up to four biological replicates are shown, with standard errors. Abbreviations: 22-OHCR, 22-hydroxycampesterol; 22-OH-3-one, 22-hydroxy-5α-ergostan-3-one; 3-epi-6-DeoxoCT, 3-epi-6-deoxocathasterone; 6-DeoxoCT, 6-deoxocathasterone; 6-DeoxoTE, 6-deoxoteasterone; 6-Deoxo3DT, 3-dehydro-6-deoxoteasterone; 6-DeoxoTY, 6-deoxotyphasterol; 6-DeoxoCS, 6-deoxocastasterone; CT, cathasterone; TE, teasterone; TY, typhasterol; CS, castasterone; BL, brassinolide; ND, not detected. (d) qRT-PCR analysis of BR biosynthetic genes in Ws-2, bri1-5, phyB-77 and phyB-77 bri1-5 seedlings grown in red light for 5 days with a photon fluence rate of 30 μmol m−2 s−1 at 22°C. Error bars represent standard errors. Values were normalized to the expression of UBQ10.
To further examine the contribution of BZR1 in phyB signaling, we generated the phyB-78 35Spro:BZR1 line and determined the ratio of dephosphorylated BZR1 (dBZR1) to phosphorylated BZR1 (pBZR1; Figure 2a), a read-out of the BR signaling status (Wang et al., 2002). The phyB-78 35Spro:BZR1 line exhibited a long hypocotyl phenotype relative to 35Spro:BZR1 transgenic seedlings (Figure 2a). Furthermore, the phosphorylated form of BZR1 was reduced in the phyB-78 background (Figure 2b). Next, we examined the expression levels of genes known to be regulated by BZR1 in a light-dependent manner (Oh et al., 2012): GER1 was suppressed, and PRE5 and PRE6 were upregulated, in phyB mutants relative to the wild type (Figure 2c), suggesting that the elongated phenotype of phyB depends on the activation of BZR1 signaling.
Figure 2.
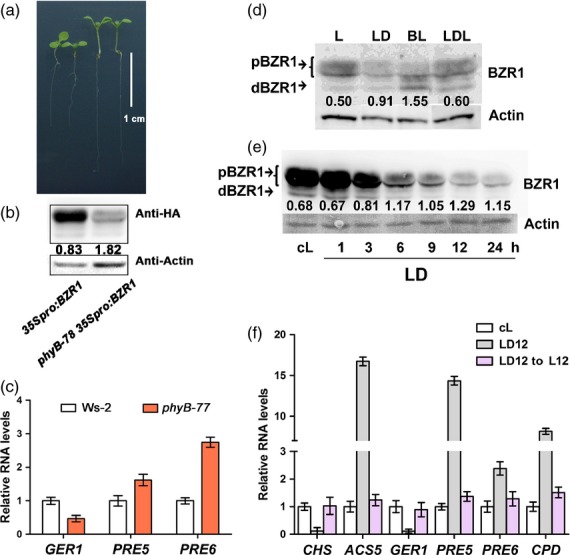
Brassinosteroid (BR) signal increases in the dark. (a) Phenotypes of 7-day-old 35Spro:BZR1-HA and phyB-78 35Spro:BZR1-HA seedlings, and the corresponding (b) protein gel blot. The numbers indicate the ratio of dBZR1 to pBZR1. (c) qRT-PCR analysis of GER1, PRE5 and PRE6 in Ws-2 and phyB-77 seedlings grown under red light for 5 days with a photon fluence rate of 80 μmol m−2 s−1 at 22°C. (d, e) Protein gel blots showing the abundance of the phosphorylated (pBZR1) and dephosphorylated (dBZR1) forms of BZR1 in different light conditions. (d) Seedlings (7 days old) expressing BZR1pro:BZR1-HA were subjected to various conditions: L, continuous light; LD, continuous light followed by transfer to dark for 6 h; BL, brassinolide treatment; LDL, transfer to darkness for 6 h and subsequently to light for 6 h with a photon fluence rate of 80 μmol m−2 s−1 at 22°C. (e) 35Spro:BZR1-HA seedlings were grown in continuous light for 7 days with a photon fluence rate of 103 μmol m−2 s−1, and were transferred to darkness for 1–24 h, as indicated. In both panels, actin was used as an equal loading control and the numbers indicate the ratio of dBZR1 to pBZR1. (f) qRT-PCR analysis of the expression of CHALCONE SYNTHASE (CHS), ACS5, GER1, PRE5, PRE6 and CPD in 10-day-old plants grown under continuous light (cL), light followed by darkness for 12 h (LD12) or light transferred to darkness for 12 h and back to light for 12 h (LD12 → L12). The photon fluence rate during light treatment was 103 μmol m−2 s−1. Error bars in (c) and (f) are standard deviations (n = 3).
phyB-77 activates BR signaling by reducing the abundance of BZR1 proteins
To identify the precise role of BZR1 in phyB-77, we sought to establish how light signaling modulates BR signaling pathways. As phyB mutants exhibits the same growth patterns as those of plants grown under shade, we performed the following experiments in total darkness or continuous white light (Figure 2d). After subjecting BZR1pro:BZR1-HA plants to various light conditions (L, light-grown control; LD, transferred to darkness for 6 h; LDL, transferred to darkness for 6 h and subsequently to light for 6 h), we found that most of the phosphorylated forms of BZR1 (pBZR1) disappeared in LD (Figure 2d), similar to the case under BL treatment (lane BL in Figure 2d), whereas the amount of protein in the dephosphorylated form (dBZR1) was similar for both LD and L (Figure 2d). The ratio of dBZR1 to pBZR1 increased upon both dark and BL treatment, indicating that BR signaling was activated by dark treatment. Conversely, when the dark-treated samples were returned to the light, pBZR1 became the dominant form (Figure 2d). Furthermore, a time-course analysis of a 35Spro:BZR1-HA line confirmed the darkness-dependent removal of BZR1 (Figure 2e). BZR1 levels gradually decreased and the dBZR1/pBZR1 ratio increased from as early as 3 h after dark treatment (Figure 2e). In support of the effect of darkness on BZR1 activity, the expression of marker genes known to be activated by BZR1, such as of ACC SYNTHASE 5 (ACS5), PRE5, PRE6 and CPD (Oh et al., 2012), was upregulated in the LD12 sample (light-grown seedlings transferred to darkness for 12 h before RNA isolation), and reverted to control levels upon return to light in the sample designated as ‘LD12 → L12’ (Figure 2f). In addition, genes known to be repressed by darkness were downregulated in LD (Figure 2f). This dark-specific removal of BZR1, preferentially the phosphorylated form, and the simultaneous regulation of known marker genes suggest that darkness activates BR signaling and hypocotyl growth by changing the abundance of BZR1.
Regulation of BZR1 stability by COP1
The dramatic removal of BZR1 upon dark treatment led us to speculate that BZR1 degradation is associated with a 26S proteasome. When light-grown seedlings were treated with both darkness and MG132, a 26S proteasome inhibitor, BZR1 noticeably accumulated (Figure 3a), suggesting that the 26S proteasome is involved in the removal of BZR1. Therefore, we next sought to identify an E3 ligase that binds to BZR1. We tested COP1, because it is known to be active in darkness (Osterlund et al., 2000; Seo et al., 2004). Compared with the Col-0 wild type (Figure 3b, lane 2), the cop1-4 mutant accumulated BZR1 even after dark treatment (lane 5), suggesting that COP1 is involved in regulating BZR1 stability. An interaction between COP1 and BZR1 was further assessed through in vivo co-immunoprecipitation (CoIP) analyses (Figure 3c). Seedlings of double transgenic lines, 35Spro:BZR1-HA 35Spro:COP1-GFP, were grown under long-day conditions, transferred to darkness and incubated for two more days. The total proteins were prepared and immunoprecipitated using anti-GFP antibody. When the precipitated proteins were separated and immunoblotted with anti-HA antibody, BZR1-HA proteins were detected (Figure 3c). CoIP data indicate that COP1 interacted with BZR1, especially pBZR1 (Figure 3c). A possible interaction between COP1 and BZR1 was further assessed through yeast two-hybrid analysis (Figure 3d). Similar to a positive control that involved the binding of BIN2 to BZR1 (He et al., 2002), COP1 bound to BZR1 (Figure 3d). Both CoIP and the yeast two-hybrid test suggest that BZR1 and COP1 bind together as part of biological signal processing mechanisms.
Figure 3.
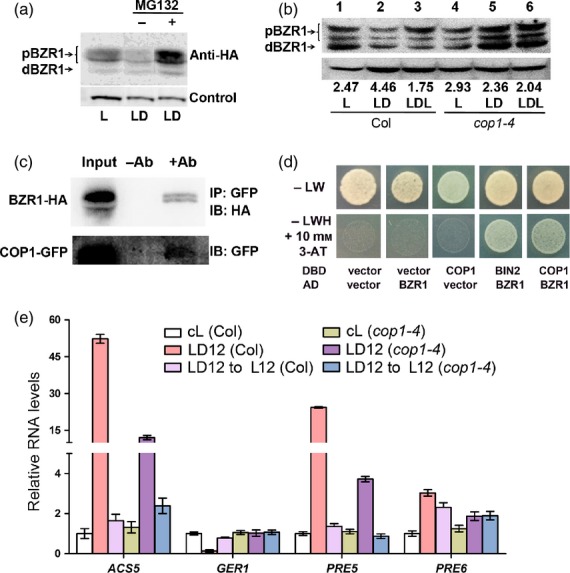
BZR1 stability is regulated by COP1 in a 26S proteasome-dependent manner. (a) Immunoblot analysis of BZR1 proteins. Seven-day-old light-grown seedlings expressing BZR1pro:BZR1-HA were treated or not with MG132 in the dark for 6 h. Actin was used as a loading control. (b) Immunoblot analysis results of the 10-day-old seedlings grown under continuous light and subsequently subjected to LD (light to dark for 12 h) or LDL (light to dark for 12 h and back to light for 12 h) growth conditions. The total protein extracted from 10-day-old seedlings was immunoblotted with anti-BZR1. Actin was used as a loading control. (c) Coimmunoprecipitation assay to test for the interaction between BZR1 and COP1 in 35Spro:BZR1-HA 35Spro:COP1-GFP Arabidopsis seedlings. Seedlings were grown under long-day conditions (with a photon fluence rate of 103 μmol m−2 s−1), and then transferred to the dark for 2 days. Total proteins were first immunoprecipitated with anti-GFP, and were subsequently immunoblotted with anti-HA and anti-GFP. (d) Yeast two hybrid analysis. –LW indicates growth media lacking leucine (L) and tryptophan (W). DBD and AD are the vectors harboring the DNA binding domain and the DNA activation domain, respectively. Vector indicates the empty vector. (e) qRT-PCR analyses of ACS5, GER1, PRE5 and PRE6 in Col-0 and cop1-4 treated with the same growth and light conditions as described in (b).
We then reasoned that the expression patterns of the marker genes would be affected in the cop1-4 mutant background. We monitored the expression of various marker genes in both wild-type and cop1-4 seedlings grown in three different conditions: continuous light (cL); continuous light-grown seedlings transferred to darkness for 12 h (LD12); and LD12 seedlings transferred back to the light for 12 h (LD12 → L12). Compared with the wild-type control, the induction of ACS5, PRE5 and PRE6 by darkness was greatly suppressed in cop1-4 mutants (Figure 3e). However, the darkness-dependent induction of these genes was canceled in the LD12 → L12 samples of both wild-type and cop1-4 backgrounds, suggesting that light-dependent repression regularly occurs in cop1-4 (Figure 3e). In support of this, we found that BZR1 accumulated in a similarly treated LDL sample of cop1-4 mutant seedlings (Figure 3b). An oppositely regulated gene, GER1, was also misregulated in cop1-4 (Figure 3e). Our CoIP results, coupled with the findings that BZR1 accumulates in cop1-4 seedlings and that gene expression patterns are altered in cop1-4, suggest that binding of COP1 to BZR1 mediates preoteasome-dependent degradation, and that the uncoupling of BZR1 degradation from darkness-triggered signaling caused misexpression of the BZR1-regulated marker genes.
Identification of genes co-regulated by both light and BR signaling
To define the list of genes that are co-regulated by BRs and light, we performed a genome-wide transcriptome analysis using total RNAs prepared from the seedlings of four genotypes, i.e. Ws-2 wild type, phyB-77, bri1-5 and phyB-77 bri1-5 (Figure 4). We constructed a heat map based on 624 genes obtained after filtering with the criteria of P < 0.01 and fold change > 1.5 in the following three comparisons: phyB-77 versus Ws-2; bri1-5 versus Ws-2; and phyB-77 bri1-5 versus Ws-2 (Figure 4a,b; Tables S1–S5). We used these comparisons because they enabled us to identify epistatic interactions. According to the expression patterns summarized in Figure 4(b), bri1-5 is epistatic to phyB-77 for both ‘bd’ (i.e.genes upregulated in phyB-77 but downregulated in the bri1-5 single and phyB-77 bri1-5 double mutant, relative to the wild type) and ‘bu’ (i.e. upregulated in the bri1-5 single and phyB-77 bri1-5 double mutant), and phyB-77 is epistatic to bri1-5 for both the ‘pu’ (i.e. upregulated in phyB-77 and the double mutant) and ‘pd’ (i.e. downregulated in phyB-77 and the double mutant, but upregulated in bri1-5) types. To validate the microarray data, we subjected the genes marked by arrowheads (Figure 4b) to quantitative RT-PCR analysis. The two genes, At5g53870 and At1g17810, encode early nodulin-like protein 1 (ENODL1) and a major intrinsic protein family water channel protein, respectively, and were upregulated in phyB-77 but were repressed in bri1-5. In contrast, At3g30180 and At5g57530 encode CYP85A2 (BR6ox2) and xyloglucan endotransglycosylase 12 (XTH12), respectively, and were found to be repressed in phyB-77 but induced in bri1-5 in both microarray and qRT-PCR analyses (Figure 4c). In addition, a Venn diagram analysis revealed 123 genes at the intersection of phyB-77, bri1-5, phyB-77 bri1-5 and the genes defined as BR-regulated BZR1 targets (BRBTs; Figure 4d; Oh et al., 2012). We postulate that the growth pattern regulated by light and BRs depends on complex interactions between these genes (Table S6).
Figure 4.
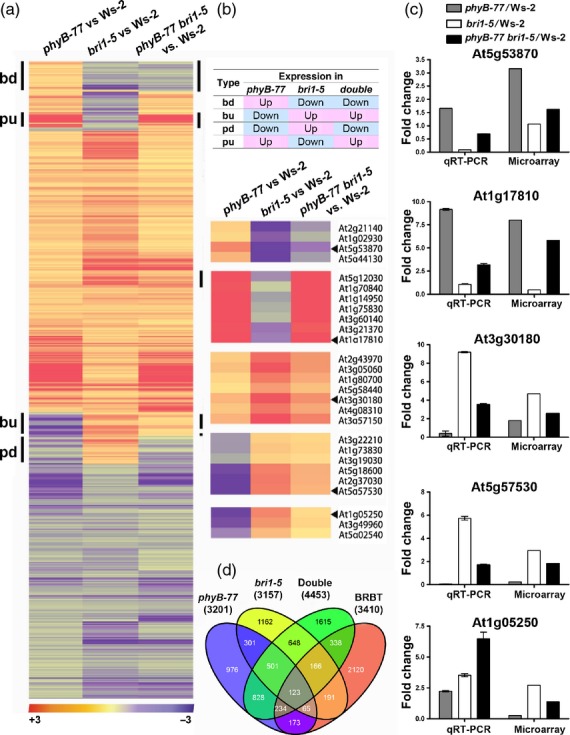
Identification of genes regulated by both light and brassinosteroids (BRs). (a, b) Heat maps of the 624 hierarchically clustered genes for which expression levels changed by more than 1.5-fold in phyB-77, bri1-5 and phyB-77 bri1-5, relative to the Ws-2 wild type. Seedlings were grown in red light for 5 days with a photon fluence rate of 30 μmol m−2 s−1 at 22°C. Genes labeled as ‘bd’ at the left of the heat map were upregulated in phyB-77 but downregulated in the bri1-5 single and phyB-77 bri1-5 double mutant, relative to the wild type. Genes downregulated in phyB-77, but upregulated in the bri1-5 single and phyB-77 bri1-5 double mutant were labeled ‘bu’. Genes designated as ‘pu’ were upregulated in phyB-77 and the double mutant, but downregulated in bri1-5, and those named ‘pd’ were downregulated in phyB-77 and the double mutant, but upregulated in bri1-5. (b) Definition of the gene clusters, and magnified view of the groups marked with vertical bars. (c) Validation of the microarray data using qRT-PCR. Error bars are the standard deviations (n = 3). (d) Venn diagram of the co-regulated genes in phyB-77, bri1-5 and phyB-77 bri1-5, relative to Ws-2, and the set of genes defined as BZR1 binding target (BRBT) genes (Oh et al., 2012).
Functional validation of the identified genes
To understand the effects of these genes on growth, the 35Spro:At5g53870-HA construct was introduced into the bri1-5 mutant background (Figure 5a,b). We found that the dwarf phenotype of bri1-5 was suppressed, such that petioles elongated and leaves expanded (Figure 5a). The expression of At5g53870 was confirmed by protein gel blot analysis (Figure 5b). Furthermore, we expressed two PEROXIDASEs (PRX2 and PRX73) that were downregulated in phyB-77 but were upregulated in bri1-5 in the antisense orientation; both PRX2 RNAi and PRX73 RNAi transgenic plants exhibited elongated hypocotyls (Figure 5c,d), and the expression of the target genes was greatly reduced (Figure 5e).
Figure 5.
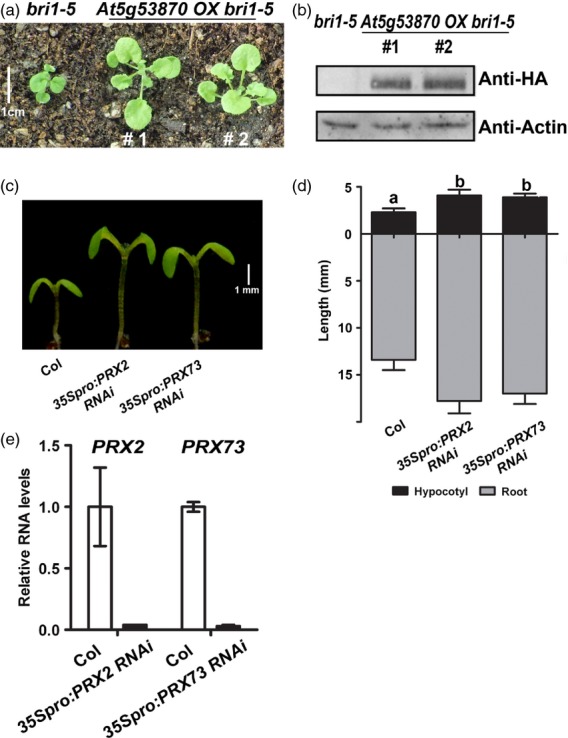
Knock-in of ENODL1 (At5g53870) and knock-down of peroxidases stimulates growth. (a, b) Phenotypes and protein levels of bri1-5 plants and two transgenic lines overexpressing At5g53870 in the bri1-5 background. (a) Comparison of 4-week-old plants exhibiting the partial suppression of bri1-5. (b) Immunoblot analysis showing At5g53870 expression in transgenic lines, compared with the bri105 control. Proteins were extracted from the aerial tissues of adult plants. (c) 35Spro:PRX2 RNAi and 35Spro:PRX73 RNAi seedlings display elongated hypocotyls. (d) Measurement of hypocotyl and root lengths. Seedlings were grown on MS medium for 7 days under long-day conditions (16-h light/8-h dark, with a photon fluence rate of 80 μmol m−2 s−1 at 22°C. Error bars are standard deviations (n = 20; P < 0.001). Different lowercase letters above each bar indicate statistically significant differences. (e) Relative RNA levels of nascent PRX2 and PRX73 in wild-type and transgenic lines expressing PRX RNAi. Error bars are standard deviations (n = 3; P < 0.001).
Formation of dimers between phosphorylated and dephosphorylated forms of BZR1
Next, we sought to explain the darkness-dependent degradation of pBZR in terms of BR signaling. We hypothesized that BZR1 proteins with different levels of phosphorylation may bind with each other as part of a regulatory mechanism. To demonstrate binding between BZR1 proteins, we transiently expressed the epitope-tagged BZR1 constructs in protoplasts and carried out a CoIP analysis (Figure 6a,b). In our transient expression system, 35Spro:BZR1-HA and 35Spro:BZR1-cMyc constructs produced phosphorylated forms of BZR1 (Figure 6a, lane 1). The 35Spro:mBZR1-HA and 35Spro:mBZR1-cMyc constructs were previously reported to have mutations of S130A and S130A/S134A, respectively, in BZR1 that reduced the levels of phosphorylation (Ryu et al., 2007). We were able to clearly discern the proteins derived from these constructs as phosphorylated (shifted toward the top of the gel; Figure 6a, lanes 1 and 4) or dephosphorylated (Figure 6b, lanes 1 and 4) forms.
Figure 6.
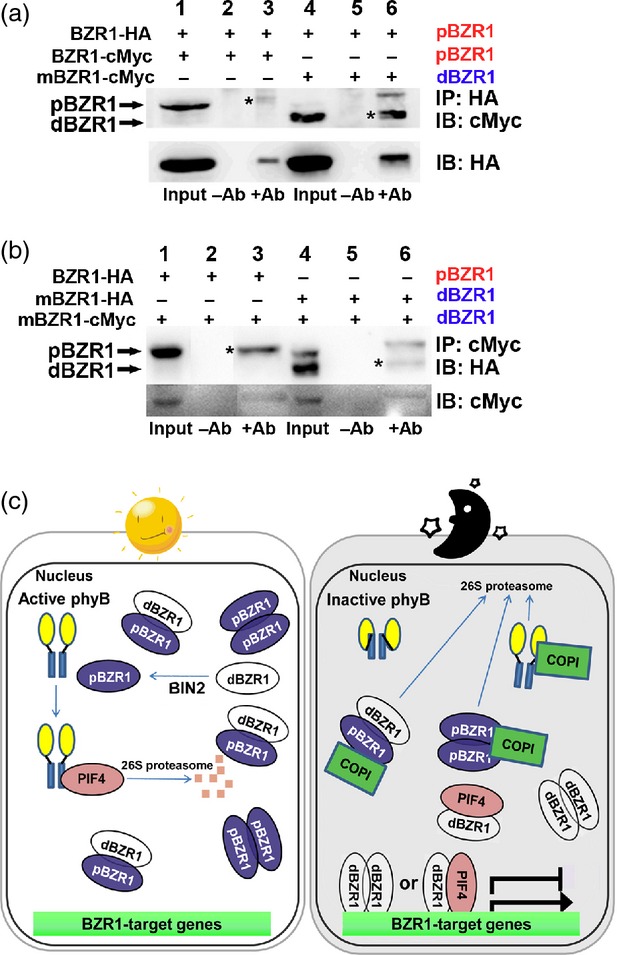
Different forms of BZR1 bind with each other as part of a regulatory mechanism. (a, b) Coimmunoprecipitation analysis showing the interaction between pBZR1 and dBZR1. Three-week-old Arabidopsis plants were used to isolate mesophyll-derived protoplasts that were transfected with 35Spro:BZR1-HA and 35Spro:BZR1-cMyc variants. Plants were grown under long-day conditions (16-h light/8-h dark, with a photon fluence rate of 80 μmol m−2 s−1 at 22°C. Protein was immunoprecipited with anti-HA (a) or anti-cMyc (b) antibody. mBZR1-HA and mBZR1-cMyc indicate BZR1S130A-HA and BZR1S130A/134A-cMyc, respectively. Protein bands with asterisks are compared for intensity. (c) Working model of the interaction between light and the brassinosteroid signaling pathways.
Lane 3 in Figure 6a shows the pBZR1-cMyc protein derived from a complex of pBZR1-cMyc and pBZR1-HA, and lane 6 indicates dBZR1-cMyc derived from the pBZR1-HA and dBZR1-cMyc complex (asterisk). This CoIP result suggests that BZR1 proteins with different levels of phosphorylation can form dimers with each other. To avoid a possible bias derived from the epitopes, and to determine the affinity of dBZR1 for dBZR1, we repeated the pull-down assay (Figure 6b). The pBZR1-HA derived from the pBZR-HA and dBZR1-cMyc complex (lane 3), and the dBZR-HA proteins from the dBZR-HA and dBZR1-cMyc complex (lane 6), were clearly detected. Interestingly, it seemed that pBZR-HA and dBZR1-cMyc (lane 3) had a greater affinity for each other than did dBZR-HA and dBZR1-cMyc (lane 6). The results of our pull-down assays suggest that BZR1 proteins can form dimers, and that pBZR1-pBZR1, pBZR1-dBZR1 and dBZR1-dBZR1 pairings are possible.
In conclusion, we propose a model that explains the roles of BZR1 in light and dark conditions. In the light (Figure 6c, left panel), the majority of BZR1 is phosphorylated by BIN2 and remains inactive in the cytoplasm (Ryu et al., 2007). Our results show that pBZR1 can trap dBZR1 to form a dimer between pBZR1 and dBZR1. In addition, PIF4 is labile in the light and less likely to form an active heterodimer with dBZR1. Because COP1 is not active in the nucleus in the light, the pools of pBZR1 and dBZR1 are maintained; however, in darkness (Figure 6c, right panel), COP1 becomes active and preferentially targets BZR1 for degradation, because pBZR1 is the major form present. The dBZR1 proteins, being freed from the complex of pBZR1 and dBZR1, can now form dimers with either dBZR1 or PIF4 to actively control downstream genes. It was previously shown that PIF4 can form a dimer with BZR1 and that the dimer controls the target genes that are regulated by both light and BRs (Oh et al., 2012).
In addition, GA is known to modulate BR signaling through controlling the stability of DELLA proteins (Bai et al., 2012). In darkness, the endogenous GA level goes up (Bae and Choi, 2008), resulting in the clearance of DELLA proteins (Itoh et al., 2003; Wang and Deng, 2011). Signaled by this increased GA level, both BZR1 and PIF proteins are released from DELLA, which enables these two proteins to form an active heterodimer and regulate the genes involved in hypocotyl growth (Feng et al., 2008; de Lucas et al., 2008; Bai et al., 2012; Jaillais and Vert, 2012). Our results fit well with this model. COP1-mediated degradation of pBZR1 should increase the possibility of dBZR1 to meet either dBZR1 or PIF4. The concerted action of GA and BR signals explains the mechanisms of hypocotyl growth under different light conditions. It is expected that future studies will reveal whether DELLA preferentially binds to pBZR1, and whether this binding affects the stability of pBZR1 protein in light.
We thus revealed a mechanism by which Arabidopsis seedlings can grow faster in darkness without a simultaneous increase in the level of bioactive BRs. Light conditions regulate the stability of BZR1 via COP1 E3 ligase to control the formation of active dimers of dBZR1-dBZR1 or dBZR1-PIF4.
EXPERIMENTAL PROCEDURES
Plant materials and growth conditions
Arabidopsis seeds were surface sterilized and plated on agar-solidified MS media. After stratification at 4°C for 3 days, seedlings were grown under long-day (16-h light/8-h dark), continuous red light or continuous white light conditions at 22°C.
To introduce 35Spro:ENODL1 (At5g53870) into bri1-5, and 35pro:COP1-GFP, 35Spro:BZR1-HA and 35Spro:PRX RNAi constructs into wild-type plants, coding sequences of each gene were cloned into pENTR/SD/D_TOPO (Invitrogen, now Life Technologies, http://www.lifetechnologies.com), and then combined with the destination vector pEarley101, pMDC83 and pB7GWIWGII in frame with epitopes, respectively. The constructs were transformed into Arabidopsis using conventional techniques.
Quantitative analysis of endogenous BRs and sterols by gas chromatography–mass spectrometry (GC-MS)
To determine the endogenous levels of both sterols and BRs, the aerial parts of 5-week-old Arabidopsis plants were harvested to yield approximately 30 g of fresh tissues, and then lyophilized at –80°C. The tissues were extracted twice with 300 ml of methanol. Deuterium-labeled internal standards were added to the extracts. Purification and quantification of sterols and BRs were performed according to a method described previously (Fujioka et al., 2002).
Coimmunoprecipitation
Previously, it was shown that site-directed substitution of Ser130 alone, or of both Ser130 and Ser134, with Ala in BZR1, designated as mBZR1-HAS130A and mBZR1-cMycS130,134A, respectively, exhibited lower levels of phosphorylation than a wild-type form (Ryu et al., 2007). These mutated versions were used to represent dephosphorylated forms. The plasmid DNA constructs were transfected into leaf mesophyll-derived protoplasts that were freshly isolated from 3–week-old wild-type plants as previously described (Hwang and Sheen, 2001). Briefly, approximately 2 × 105 protoplasts were transfected with 30 μg BZR1 and mBZR1 plasmid DNA. Transfected protoplasts were incubated for 6 h at room temperature and homogenized in immunoprecipitation buffer containing 50 mm Tris–HCl (pH 7.5), 100 mm NaCl, 2 mm DTT, 0.1% Triton X-100, and full strength (1X) protease inhibitor cocktail (GenDEPOT, http://www.gendepot.com). The supernatant was incubated for 3 h at 4°C with agarose-conjugated anti-HA (Roche, http://www.roche.com) and then washed three times with dH2O. Then, 30 μl of denaturated proteins was subjected to SDS-PAGE on a 10% gel. After transfer to polyvinylidene difluoride membranes, the blot was detected with anti-cMyc (Cell Signaling Technology, http://www.cellsignal.com) or anti-HA (Roche) diluted at 1/2000. The immunoblot was washed with phosphate-buffered saline buffer and incubated with the corresponding anti-horseradish peroxidase (HRP) antibodies for visualization of the protein bands (GenDEPOT).
Yeast two-hybrid assays
Full-length coding sequences of BZR1, BIN2 and COP1 were cloned into the low-copy yeast expression vectors pDEST22 (Gal4 AD) and pDEST32 (Gal4 BD). To test protein interactions, the corresponding plasmids were co-transformed into the yeast MaV203 strain. Successfully transformed colonies were identified on yeast synthetic drop-out media that lacked Leu and Trp (Clontech, http://www.clontech.com). At 3 days after transformation, yeast colonies were grown in selective –Leu, –Trp liquid media for 24 h, and the cell density was adjusted to OD600 = 0.1. An ∼10-μl volume of cell suspension was pipetted onto yeast synthetic drop-out media lacking His, Leu and Trp, and supplemented with 10 mM 3-amino-1,2,4-triazole (3-AT; Sigma-Aldrich, http://www.sigmaaldrich.com). Plates were incubated at 28°C for 2–4 days. The empty vectors pDEST22 and pDEST32 were co-transformed as negative controls.
Protein extraction and immunoblot analysis
Total proteins were extracted from the tissues of 7-day-old seedlings grown on agar-solidified media using protein extraction buffer [50 mm Tris–Cl (pH 7.5), 100 mm NaCl, 10 mm MgCl2, 1 mm EDTA, 10% glycerol, 1 mm DTT, 1 mm phenylmethylsulfonyl fluoride and 1X protease inhibitor cocktail (GenDEPOT)]. Proteins were equally loaded on an 8 or 10% SDS–polyacrylamide gel and transferred to a polyvinylidene difluoride membrane. The antibodies used (at 1:3000) were anti-actin (MP Biomedicals, http://www.mpbio.com) and anti-BZR1 (generated in our lab). Following three washings with Tris-buffered saline Tween-20 (TBST) buffer, membranes were incubated with HRP-linked secondary antibodies (GenDEPOT). The membranes were then washed again with TBST, and antibodies were visualized with a chemiluminescence method (Amersham Bioscience, now GE Healthcare, http://www.gelifesciences.com). Protein gel blot analyses were performed multiple times, and the data shown are representative blots.
Microarray and quantitative RT-PCR analysis
Microarray experiments were performed using an Affymetrix ATH1 GeneChip system. Seedlings of four genotypes, Ws-2 wild type, bri1-5, phyB-77 and bri1-5 phyB-77, were grown for 7 days on 1X MS agar-solidified media under continuous red light. Biological samples were analyzed in triplicate and on three chips per genotype. The microarray analysis generated 12 different CEL files. All data generated during this experiment were submitted to the Gene Expression Omnibus (GEO) (http://www.ncbi.nlm.nih.gov/projects/geo/) under accession number GSE46456. Statistical analyses of the data were performed according to methods described previously (Chung et al., 2011).
For validation of the microarray data, 7-day-old light-grown seedlings were ground to a fine powder in liquid nitrogen. Total RNA was prepared using Trizol (Sigma-Aldrich). Two micrograms of total RNA was reverse-transcribed (Fermentas, now Thermo Scientific, http://www.thermoscientificbio.com/fermentas), and equal quantities of reverse-transcribed products were used for the PCR reactions. Each template RNA was normalized using the UBQ10 (AT4G05320) or TUB2 (AT5G62690) genes as loading controls. Quantitative RT-PCR was performed in 96-well plates (Applied Biosystems, http://www.appliedbiosystems.com) using a commercial qPCR kit (KapaBiosystems, http://www.kapabiosystems.com) in a volume of 20 μl. The reactions were performed in triplicate. Samples were quantified by generating standard curves using the results obtained with serially diluted control DNA. The relative mRNA levels represent fold change over the control. Each cDNA was normalized using UBQ10 or TUB2. The oligonucleotide sequences used in the qRT-PCR are listed in Table S7.
Acknowledgments
We would like to thank J. Chory for the gift of bzr1-D and bes1-GFP seeds, and S. Takatsuto for supplying deuterium-labeled internal standards. We are also indebted to the Choe lab members, including Y.C., P.M.M., M.Y.K., W.S.K., and O.S.L., for their technical assistance. This research was supported, in part, by grants from the Next-Generation BioGreen 21 Program (Plant Molecular Breeding Center, project no. PJ008051) and Cooperative Research Program for Agriculture Science & Technology Development (project no. PJ906910), Rural Development Administration (to C.S.); the BK21 Research Fellowships (to C.C.) funded by the Ministry of Education, Science and Technology, Republic of Korea; and a Grant-in-Aid for Scientific Research (B) from the Ministry of Education, Culture, Sports, Science and Technology of Japan (19380069 and 23380066 to S.F.).
SUPPORTING INFORMATION
Additional Supporting Information may be found in the online version of this article.
Complete data of the microarray analysis using the RNAs prepared from Ws–2 wild type, phyB-77, bri1-5 and phyB-77 bri1-5 seedlings grown for 7 days under continuous red light.
Expression data for 3201 genes, with a comparison of phyB-77 versus Ws-2 (P < 0.01).
Expression data for 3157 genes, with a comparison of bri1-5 versus Ws-2 (P < 0.01).
Expression data for 4453 genes, with a comparison of phyB-77 bri1-5 versus Ws-2 (p < 0.01).
Expression data for 624 genes, with P < 0.01 at all three comparisons of phyB-77 versus Ws-2, bri1-5 versus Ws-2 and phyB-77 bri1-5 versus Ws-2.
Expression data for the genes at each intersection in the Venn diagram.
List of primers used in quantitative RT-PCR and cloning.
REFERENCES
- Alabadi D, Gallego-Bartolome J, Orlando L, et al. Gibberellins modulate light signaling pathways to prevent Arabidopsis seedling de-etiolation in darkness. Plant J. 2008;53:324–335. doi: 10.1111/j.1365-313X.2007.03346.x. [DOI] [PubMed] [Google Scholar]
- Bae G, Choi G. Decoding of light signals by plant phytochromes and their interacting proteins. Annu. Rev. Plant Biol. 2008;59:281–311. doi: 10.1146/annurev.arplant.59.032607.092859. [DOI] [PubMed] [Google Scholar]
- Bai MY, Shang JX, Oh E, Fan M, Bai Y, Zentella R, Sun TP, Wang ZY. Brassinosteroid, gibberellin and phytochrome impinge on a common transcription module in Arabidopsis. Nat. Cell Biol. 2012;14:810–817. doi: 10.1038/ncb2546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bou-Torrent J, Roig-Villanova I, Galstyan A, Martinez-Garcia JF. PAR1 and PAR2 integrate shade and hormone transcriptional networks. Plant Signal. Behav. 2008;3:453–454. doi: 10.4161/psb.3.7.5599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carabelli M, Morelli G, Whitelam G, Ruberti I. Twilight-zone and canopy shade induction of the Athb-2 homeobox gene in green plants. Proc. Natl Acad. Sci. USA. 1996;93:3530–3535. doi: 10.1073/pnas.93.8.3530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choe S, Dilkes BP, Fujioka S, Takatsuto S, Sakurai A, Feldmann KA. The DWF4 gene of Arabidopsis encodes a cytochrome P450 that mediates multiple 22α-hydroxylation steps in brassinosteroid biosynthesis. Plant Cell. 1998;10:231–243. doi: 10.1105/tpc.10.2.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choe S, Dilkes BP, Gregory BD, et al. The Arabidopsis dwarf1 mutant is defective in the conversion of 24-methylenecholesterol to campesterol in brassinosteroid biosynthesis. Plant Physiol. 1999a;119:897–907. doi: 10.1104/pp.119.3.897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choe S, Noguchi T, Fujioka S, et al. The Arabidopsis dwf7/ste1 mutant is defective in the delta7 sterol C-5 desaturation step leading to brassinosteroid biosynthesis. Plant Cell. 1999b;11:207–221. [PMC free article] [PubMed] [Google Scholar]
- Choe S, Tanaka A, Noguchi T, Fujioka S, Takatsuto S, Ross AS, Tax FE, Yoshida S, Feldmann KA. Lesions in the sterol Δ7 reductase gene of Arabidopsis cause dwarfism due to a block in brassinosteroid biosynthesis. Plant J. 2000;21:431–443. doi: 10.1046/j.1365-313x.2000.00693.x. [DOI] [PubMed] [Google Scholar]
- Choe S, Fujioka S, Noguchi T, Takatsuto S, Yoshida S, Feldmann KA. Overexpression of DWARF4 in the brassinosteroid biosynthetic pathway results in increased vegetative growth and seed yield in Arabidopsis. Plant J. 2001;26:573–582. doi: 10.1046/j.1365-313x.2001.01055.x. [DOI] [PubMed] [Google Scholar]
- Choe S, Schmitz RJ, Fujioka S, Takatsuto S, Lee MO, Yoshida S, Feldmann KA, Tax FE. Arabidopsis brassinosteroid-insensitive dwarf12 mutants are semidominant and defective in a glycogen synthase kinase 3β-like kinase. Plant Physiol. 2002;130:1506–1515. doi: 10.1104/pp.010496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung Y, Maharjan PM, Lee O, et al. Auxin stimulates DWARF4 expression and brassinosteroid biosynthesis in Arabidopsis. Plant J. 2011;66:564–578. doi: 10.1111/j.1365-313X.2011.04513.x. [DOI] [PubMed] [Google Scholar]
- Chung Y, Choe V, Fujioka S, Takatsuto S, Han M, Jeon JS, Park YI, Lee KO, Choe S. Constitutive activation of brassinosteroid signaling in the Arabidopsis elongated-D/bak1 mutant. Plant Mol. Biol. 2012;80:489–501. doi: 10.1007/s11103-012-9963-5. [DOI] [PubMed] [Google Scholar]
- Clouse SD, Langford M, McMorris TC. A brassinosteroid-insensitive mutant in Arabidopsis thaliana exhibits multiple defects in growth and development. Plant Physiol. 1996;111:671–678. doi: 10.1104/pp.111.3.671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng S, Martinez C, Gusmaroli G, et al. Coordinated regulation of Arabidopsis thaliana development by light and gibberellins. Nature. 2008;451:475–479. doi: 10.1038/nature06448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin KA, Whitelam GC. Phytochromes and shade-avoidance responses in plants. Ann. Bot. (Lond) 2005;96:169–175. doi: 10.1093/aob/mci165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujioka S, Takatsuto S, Yoshida S. An early C-22 oxidation branch in the brassinosteroid biosynthetic pathway. Plant Physiol. 2002;130:930–939. doi: 10.1104/pp.008722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gampala SS, Kim TW, He JX, et al. An essential role for 14-3-3 proteins in brassinosteroid signal transduction in Arabidopsis. Dev. Cell. 2007;13:177–189. doi: 10.1016/j.devcel.2007.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He JX, Gendron JM, Yang Y, Li J, Wang ZY. The GSK3-like kinase BIN2 phosphorylates and destabilizes BZR1, a positive regulator of the brassinosteroid signaling pathway in Arabidopsis. Proc. Natl Acad. Sci. USA. 2002;99:10185–10190. doi: 10.1073/pnas.152342599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang I, Sheen J. Two-component circuitry in Arabidopsis cytokinin signal transduction. Nature. 2001;413:383–389. doi: 10.1038/35096500. [DOI] [PubMed] [Google Scholar]
- Itoh H, Matsuoka M, Steber CM. A role for the ubiquitin-26S-proteasome pathway in gibberellin signaling. Trend Plant Sci. 2003;8:492–497. doi: 10.1016/j.tplants.2003.08.002. [DOI] [PubMed] [Google Scholar]
- Jaillais Y, Vert G. Brassinosteroids, gibberellins and light-mediated signalling are the three-way controls of plant sprouting. Nat. Cell Biol. 2012;14:788–790. doi: 10.1038/ncb2551. [DOI] [PubMed] [Google Scholar]
- Kim J, Yi H, Choi G, Shin B, Song PS, Choi G. Functional characterization of phytochrome interacting factor 3 in phytochrome-mediated light signal transduction. Plant Cell. 2003;15:2399–2407. doi: 10.1105/tpc.014498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim B, Fujioka S, Kwon M, Jeon J, Choe S. Arabidopsis brassinosteroid-overproducing gulliver3-D/dwarf4-D mutants exhibit altered responses to jasmonic acid and pathogen. Plant Cell Rep. 2013;32:1139–1149. doi: 10.1007/s00299-012-1381-2. [DOI] [PubMed] [Google Scholar]
- Kim B, Kwon M, Jeon J, Schulz B, Corvalán C, Jeong YJ, Choe S. The Arabidopsis gulliver2/phyB Mutant Exhibits Reduced Sensitivity to Brassinazole. J. Plant. Biol. 2014;57:20–27. [Google Scholar]
- Kozuka T, Horiguchi G, Kim GT, Ohgishi M, Sakai T, Tsukaya H. The different growth responses of the Arabidopsis thaliana leaf blade and the petiole during shade avoidance are regulated by photoreceptors and sugar. Plant Cell Physiol. 2005;46:213–223. doi: 10.1093/pcp/pci016. [DOI] [PubMed] [Google Scholar]
- Leivar P, Quail PH. PIFs: pivotal components in a cellular signaling hub. Trends Plant Sci. 2011;16:19–28. doi: 10.1016/j.tplants.2010.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Nam KH. Regulation of brassinosteroid signaling by a GSK3/SHAGGY-like kinase. Science. 2002;295:1299–1301. doi: 10.1126/science.1065769. [DOI] [PubMed] [Google Scholar]
- Li J, Nagpal P, Vitart V, McMorris TC, Chory J. A role for brassinosteroids in light-dependent development of Arabidopsis. Science. 1996;272:398–401. doi: 10.1126/science.272.5260.398. [DOI] [PubMed] [Google Scholar]
- Li J, Nam KH, Vafeados D, Chory J. BIN2, a new brassinosteroid-insensitive locus in Arabidopsis. Plant Physiol. 2001;127:14–22. doi: 10.1104/pp.127.1.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorrain S, Allen T, Duek PD, Whitelam GC, Fankhauser C. Phytochrome-mediated inhibition of shade avoidance involves degradation of growth-promoting bHLH transcription factors. Plant J. 2008;53:312–323. doi: 10.1111/j.1365-313X.2007.03341.x. [DOI] [PubMed] [Google Scholar]
- de Lucas M, Daviere JM, Rodriguez-Falcon M, Pontin M, Iglesias-Pedraz JM, Lorrain S, Fankhauser C, Blazquez MA, Titarenko E, Prat S. A molecular framework for light and gibberellin control of cell elongation. Nature. 2008;451:480–484. doi: 10.1038/nature06520. [DOI] [PubMed] [Google Scholar]
- Luccioni LG, Oliverio KA, Yanovsky MJ, Boccalandro HE, Casal JJ. Brassinosteroid mutants uncover fine tuning of phytochrome signaling. Plant Physiol. 2002;128:173–181. [PMC free article] [PubMed] [Google Scholar]
- Morelli G, Ruberti I. Light and shade in the photocontrol of Arabidopsis growth. Trends Plant Sci. 2002;7:399–404. doi: 10.1016/s1360-1385(02)02314-2. [DOI] [PubMed] [Google Scholar]
- Noguchi T, Fujioka S, Choe S, Takatsuto S, Yoshida S, Yuan H, Feldmann KA, Tax FE. Brassinosteroid-insensitive dwarf mutants of Arabidopsis accumulate brassinosteroids. Plant Physiol. 1999;121:743–752. doi: 10.1104/pp.121.3.743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh E, Zhu JY, Wang ZY. Interaction between BZR1 and PIF4 integrates brassinosteroid and environmental responses. Nat. Cell Biol. 2012;14:802–809. doi: 10.1038/ncb2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osterlund MT, Wei N, Deng XW. The roles of photoreceptor systems and the COP1-targeted destabilization of HY5 in light control of Arabidopsis seedling development. Plant Physiol. 2000;124:1520–1524. doi: 10.1104/pp.124.4.1520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Perez JM, Ponce MR, Micol JL. The UCU1 Arabidopsis gene encodes a SHAGGY/GSK3-like kinase required for cell expansion along the proximodistal axis. Dev. Biol. 2002;242:161–173. doi: 10.1006/dbio.2001.0543. [DOI] [PubMed] [Google Scholar]
- Pierik R, Cuppens ML, Voesenek LA, Visser EJ. Interactions between ethylene and gibberellins in phytochrome-mediated shade avoidance responses in tobacco. Plant Physiol. 2004;136:2928–2936. doi: 10.1104/pp.104.045120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu H, Kim K, Cho H, Park J, Choe S, Hwang I. Nucleocytoplasmic shuttling of BZR1mediated by phosphorylation is essential in Arabidopsis brassinosteroid signaling. Plant Cell. 2007;17:2749–2762. doi: 10.1105/tpc.107.053728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo HS, Watanabe E, Tokutomi S, Nagatani A, Chua NH. Photoreceptor ubiquitination by COP1 E3 ligase desensitizes phytochrome A signaling. Genes Dev. 2004;18:617–622. doi: 10.1101/gad.1187804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin J, Kim K, Kang H, Zulfugarov IS, Bae G, Lee CH, Lee D, Choi G. Phytochromes promote seedling light responses by inhibiting four negatively-acting phytochrome-interacting factors. Proc. Natl Acad. Sci. USA. 2009;106:7660–7665. doi: 10.1073/pnas.0812219106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorin C, Salla-Martret M, Bou-Torrent J, Roig-Villanova I, Martinez-Garcia JF. ATHB4, a regulator of shade avoidance, modulates hormone response in Arabidopsis seedlings. Plant J. 2009;59:266–277. doi: 10.1111/j.1365-313X.2009.03866.x. [DOI] [PubMed] [Google Scholar]
- Stavang JA, Gallego-Bartolome J, Gomez MD, Yoshida S, Asami T, Olsen JE, Garcia-Martinez JL, Alabadi D, Blazquez MA. Hormonal regulation of temperature-induced growth in Arabidopsis. Plant J. 2009;60:589–601. doi: 10.1111/j.1365-313X.2009.03983.x. [DOI] [PubMed] [Google Scholar]
- Steindler C, Matteucci A, Sessa G, Weimar T, Ohgishi M, Aoyama T, Morelli G, Ruberti I. Shade avoidance responses are mediated by the ATHB-2 HD-zip protein, a negative regulator of gene expression. Development. 1999;126:4235–4245. doi: 10.1242/dev.126.19.4235. [DOI] [PubMed] [Google Scholar]
- Szekeres M, Nemeth K, Koncz-Kalman Z, Mathur J, Kauschmann A, Altmann T, Redei GP, Nagy F, Schell J, Koncz C. Brassinosteroids rescue the deficiency of CYP90, a cytochrome P450, controlling cell elongation and de-etiolation in Arabidopsis. Cell. 1996;85:171–182. doi: 10.1016/s0092-8674(00)81094-6. [DOI] [PubMed] [Google Scholar]
- Tao Y, Ferrer JL, Ljung K, et al. Rapid synthesis of auxin via a new tryptophan-dependent pathway is required for shade avoidance in plants. Cell. 2008;133:164–176. doi: 10.1016/j.cell.2008.01.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandenbussche F, Pierik R, Millenaar FF, Voesenek LA, Van Der Straeten D. Reaching out of the shade. Curr. Opin. Plant Biol. 2005;8:462–468. doi: 10.1016/j.pbi.2005.07.007. [DOI] [PubMed] [Google Scholar]
- Vert G, Chory J. Downstream nuclear events in brassinosteroid signalling. Nature. 2006;441:96–100. doi: 10.1038/nature04681. [DOI] [PubMed] [Google Scholar]
- Wang F, Deng XW. Plant ubiquitin-proteasome pathway and its role in gibberellin signaling. Cell Res. 2011;21:1286–1294. doi: 10.1038/cr.2011.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ZY, Nakano T, Gendron J, et al. Nuclear-localized BZR1 mediates brassinosteroid-induced growth and feedback suppression of brassinosteroid biosynthesis. Dev. Cell. 2002;2:505–513. doi: 10.1016/s1534-5807(02)00153-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Complete data of the microarray analysis using the RNAs prepared from Ws–2 wild type, phyB-77, bri1-5 and phyB-77 bri1-5 seedlings grown for 7 days under continuous red light.
Expression data for 3201 genes, with a comparison of phyB-77 versus Ws-2 (P < 0.01).
Expression data for 3157 genes, with a comparison of bri1-5 versus Ws-2 (P < 0.01).
Expression data for 4453 genes, with a comparison of phyB-77 bri1-5 versus Ws-2 (p < 0.01).
Expression data for 624 genes, with P < 0.01 at all three comparisons of phyB-77 versus Ws-2, bri1-5 versus Ws-2 and phyB-77 bri1-5 versus Ws-2.
Expression data for the genes at each intersection in the Venn diagram.
List of primers used in quantitative RT-PCR and cloning.