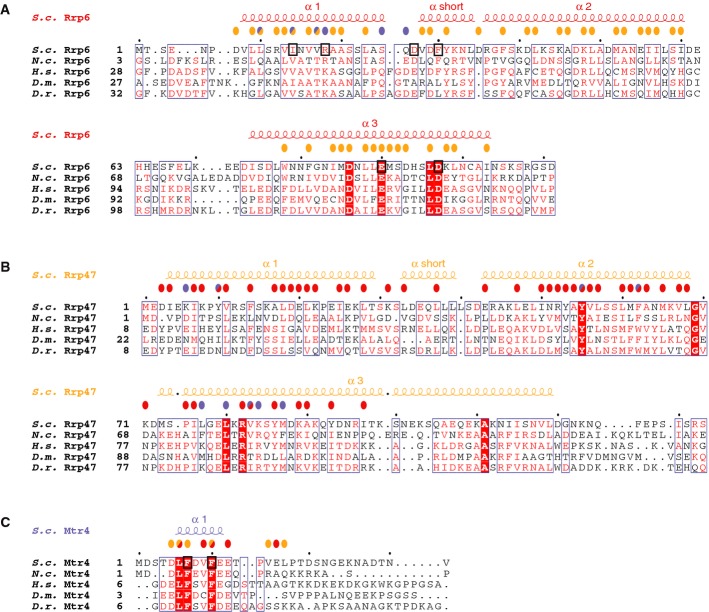
Figure 4. Structure-based sequence alignments of Rrp6N, Rrp47N and Mtr4N.
- A–C The alignments of Rrp6 (A), Rrp47 (B) and Mtr4 (C) include orthologues from the representative species Saccharomyces cerevisiae (S.c.), Neurospora crassa (N.c.), Homo sapiens (H.s.), Drosophila melanogaster (D.m.) and Danio rerio (D.r.), based on a comprehensive alignment. The secondary structure elements are shown above the sequences. Conserved residues are highlighted in color. Colored circles above the sequences identify residues involved in the interaction with Rrp6 (red circles), with Rrp47 (orange circles) and with Mtr4 (blue circles). Circles of two colors indicate residues involved in interactions with two partners in the ternary complex. Residues targeted for mutagenesis are highlighted with a black square.