Figure 7. Structure-based mutations in the Rrp6 N-terminal domain result in 5.8S rRNA processing defects in vivo.
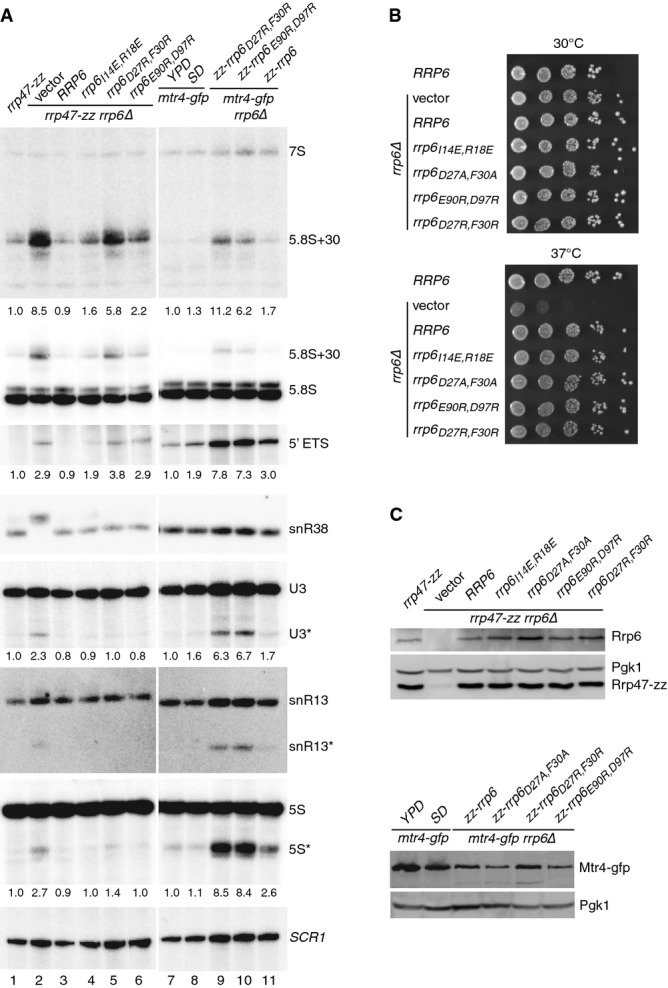
- Northern blot analyses of RNA from rrp6, mtr4-gfp and mtr4-gfp rrp6 double mutants. Strains were grown in selective minimal medium, unless indicated otherwise. Consecutive hybridizations of a single blot are shown; panels shown for lanes 1–6 and 7–11 are juxtaposed from a single image. The major RNAs detected by each probe are indicated on the right. Asterisks indicate truncated RNA fragments of U3, snR13 and 5S. The amount of ‘5.8S+30’ RNA, the 5′ ETS fragment and the major U3 and 5S degradation fragments in each mutant (indicated beneath the appropriate panel) is expressed relative to the rrp47-zz strain in lanes 1–6 and to the mtr4-gfp strain during growth in YPD in lanes 7–11.
- Spot growth assays of rrp6 mutants. Serial dilutions of pre-cultures were spotted onto selective minimal medium plates and incubated at 30 or 37°C. The plates were photographed after 3 days.
- Western analyses of rrp6 mutants. (Upper panels) Expression levels of Rrp6 and the Rrp47-zz fusion protein in the rrp6 mutants. (Lower panels) Mtr4–gfp fusion protein expression levels in the mtr4-gfp rrp6 double mutants. Pgk1 levels were analyzed as a loading control in each case.
