Figure 5. Framework for the interaction of DNA with Mre11–Rad50 functional states.
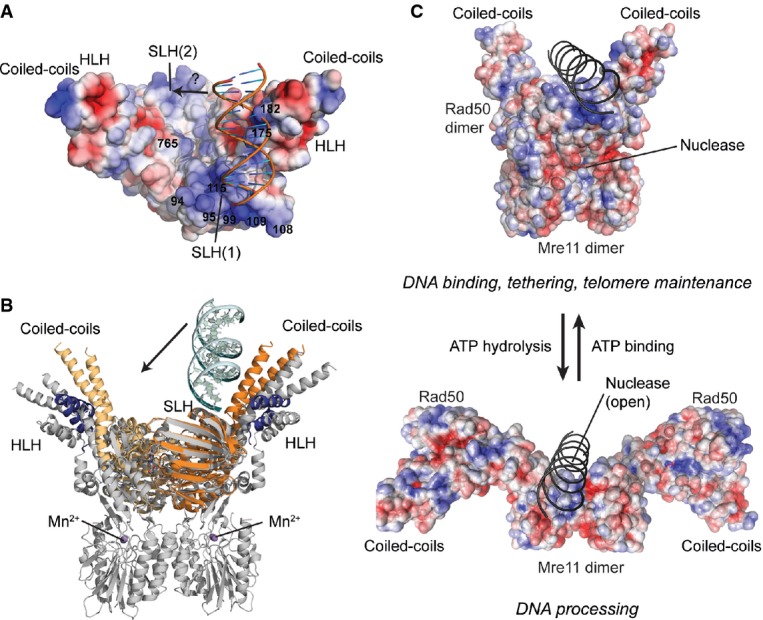
- Surface representation of tmRad50NBD–Mre11HLH with mapped electrostatic potential (blue: positive; red: negative). DNA is shown as cartoon. The SLH motifs and the surface in the groove between the coiled-coil domains carry a strong positively charged surface potential. A moderate shift of the bound DNA would enable additional contacts to the second SLH motif (see text).
- Superposition of the tmRad50NBD–Mre11HLH–DNA (color code of Fig1) and tmRad50NBD–Mre11FL (gray; PDB code 3THO) models. DNA binds to Rad50 on the opposite side of the Mre11 nuclease dimer and does not sterically compete with Mre11 binding.
- Model of DNA binding and its implication for the functions of the MRN complex. Top: the ATP-bound ‘closed’ conformation with DNA bound to Rad50 is implicated for DNA binding, tethering, and telomere maintenance functions. Upon ATP hydrolysis, Rad50 NBDs move away from each other, exposing the Mre11 active sites to allow for DNA processing.
