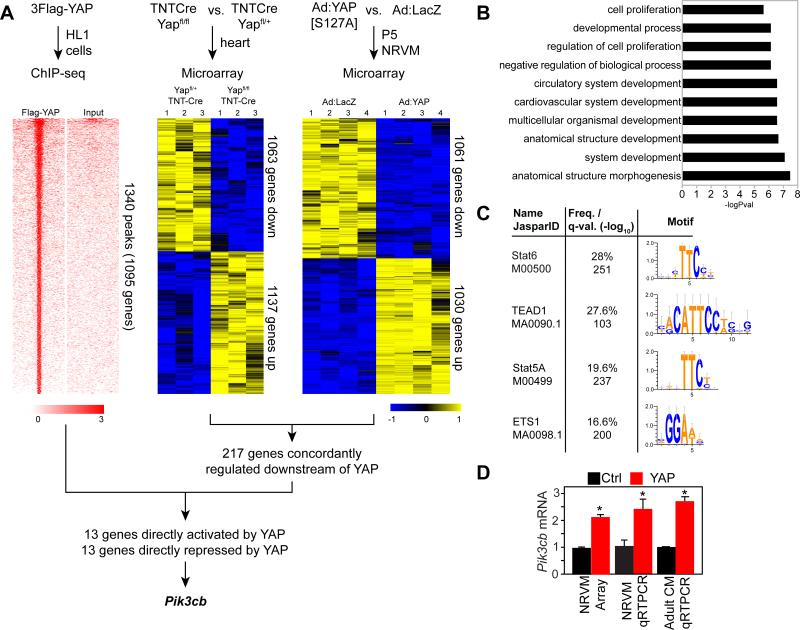
Figure 1. Genome-wide screen to identify genes directly regulated by YAP in cardiomyocytes.
A. Strategy to identify genes directly bound by YAP and differentially expressed in YAP gain- and loss-of-function. ChIP-seq was performed using Flag-tagged YAP in HL1 cells. Tag heatmap shows signal intensity in ChIP and input sample in a 4 kb window centered on YAP peaks. Differential gene expression in YAP loss-of-function and gain-of-function were determined by microarray analysis of E12.5 hearts with cardiomyocyte-restricted YAP knockout, and NRVMs that overexpressed activated YAP. Heatmap shows the row-scaled expression z-score. B. Gene Ontology term analysis of YAP- bound genes. C. Motif analysis of the top 1000 YAP bound regions. D.YAP overexpression induced upregulation of Pik3cb. *, P<0.05.