Figure 1. In vitro splicing of XBP1 mRNA and subcellular localization of RTCB and archease.
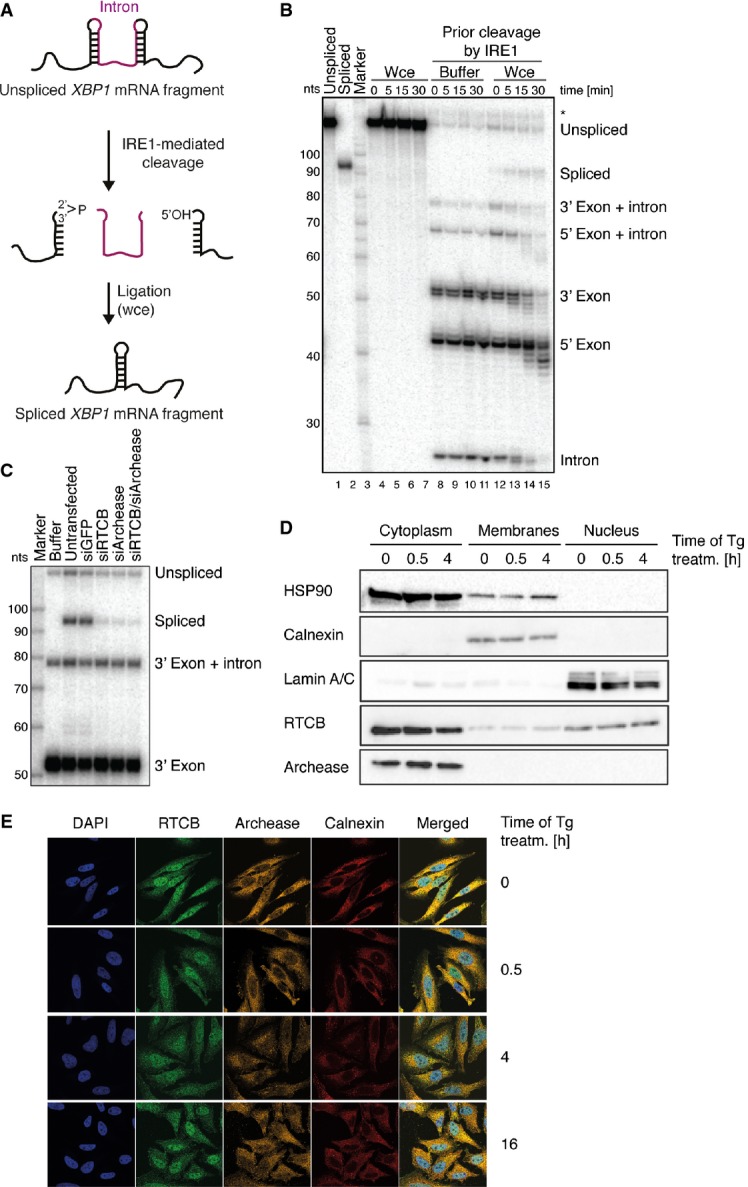
- Schematic representation of the in vitro assay to monitor XBP1 mRNA splicing. A radiolabelled human XBP1 transcript encompassing the intron is pre-cleaved with recombinant, constitutively active IRE1 to form RNA fragments mimicking XBP1 mRNA exon halves. Subsequent incubation with HeLa whole-cell extracts provides the ligation activity required to convert these fragments into a single, longer species representing the spliced form of XBP1 mRNA.
- An internally labeled fragment of XBP1 mRNA including the intron (lane 1) was incubated with HeLa whole-cell extracts (Wce, lanes 4–7) or pre-cleaved with recombinant IRE1 endonuclease and afterward supplemented with buffer (lanes 8–11) or Wce (lanes 12–15) for the indicated time periods. After addition of Wce, cleaved XBP1 mRNA fragments were efficiently converted into the spliced form XBP1 mRNA (compare to lane 2). A nucleotide (nt) size marker is shown in lane 3. An unspecific band is marked with an asterisk.
- HeLa cells were transfected with control siRNA (siGFP) or siRNAs against RTCB, archease or both and harvested 3 days post-transfection. Whole-cell extracts were incubated with a 3′ end-labeled XBP1 mRNA pre-cleaved by recombinant IRE1 for 15 min.
- Subcellular localization of RTCB and archease assessed by Western blot analysis of fractions obtained after subcellular fractionation of HeLa cells treated with 300 nM thapsigargin (Tg) for the indicated time periods. HSP90 (cytoplasm), calnexin (membranes) and lamin A/C (nucleus) were used as marker proteins for the individual fractions collected (n = 5).
- Subcellular localization of RTCB and archease visualized by immunofluorescence staining of HeLa cells treated with 300 nM Tg for the indicated time periods. The nucleus is visualized by DAPI staining. Calnexin staining is used to mark the ER membrane (n = 4).
