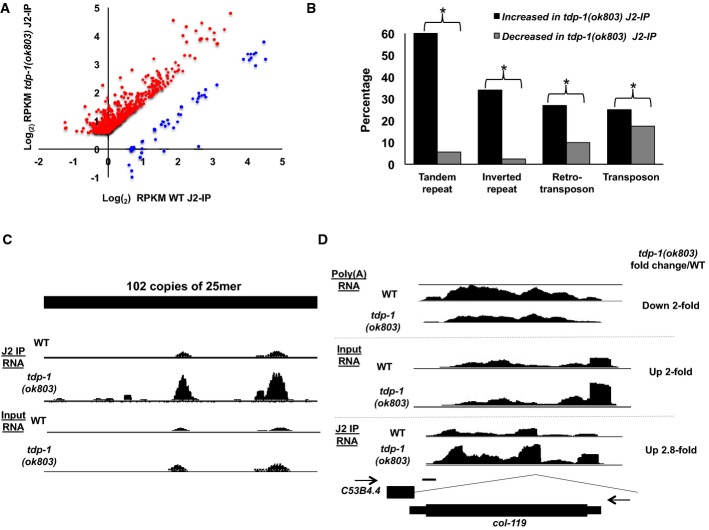
Figure 5. Double-stranded RNA transcripts are preferentially recovered in tdp-1(ok803) mutant extract.
- Graph comparing the log2 RPKM (normalized to input) of tdp-1(ok803) J2-IP (y-axis) versus wild-type J2-IP (x-axis) for all genes significantly increased (red dots) and decreased (blue dots) in representation (P < 0.05, FDR < 0.1 for all changes). Plot derived from data shown in Supplementary Fig S10 using three independent biological replicates of wild-type and tdp-1(ok803) J2-IP.
- The percentage (x-axis) of all expressed repetitive elements (type shown on y-axis) that are significantly increased (black bars) and decreased (gray bars) in tdp-1 mutant J2-IP compared to wild-type J2-IP (P < 0.05 for all changes). *P < 1 × 10−10 chi-square test. Plot derived from data shown in Supplementary Fig S12.
- A representative example of a tandem repeat region that is increased for dsRNA structure/stability in tdp-1(ok803) J2-IP. Coverage tracks from both the J2-IP RNA-seq and input RNA-seq are shown; region displayed: chromosome I: 10,130,500–10,133,500.
- Coverage tracks of col-119 gene (transcribed antisense with the intron of expressed gene C53B4.4) in poly(A) RNA-seq (top), input (total) RNA-seq (middle) and J2-IP RNA-seq (bottom) (note: scale of coverage tracks for J2-IP is normalized to input for display purposes). Gene models for both genes are shown below coverage tracks, the direction of transcription is indicated by black arrows). Scale bars (black line), 200 bp. See also Supplementary Fig S11.