Abstract
A symbiotic mutant of Lotus japonicus, called sunergos1-1 (suner1-1), originated from a har1-1 suppressor screen. suner1-1 supports epidermal infection by Mesorhizobium loti and initiates cell divisions for organogenesis of nodule primordia. However, these processes appear to be temporarily stalled early during symbiotic interaction, leading to a low nodule number phenotype. This defect is ephemeral and near wild-type nodule numbers are reached by suner1-1 at a later point after infection. Using an approach that combined map-based cloning and next-generation sequencing we have identified the causative mutation and show that the suner1-1 phenotype is determined by a weak recessive allele, with the corresponding wild-type SUNER1 locus encoding a predicted subunit A of a DNA topoisomerase VI. Our data suggest that at least one function of SUNER1 during symbiosis is to participate in endoreduplication, which is an essential step during normal differentiation of functional, nitrogen-fixing nodules.
Keywords: legumes, Lotus japonicus, symbiosis, topoisomerase, endocycle
Introduction
Endosymbiosis is a relationship in which members of one species live inside another species; nitrogen-fixing nodular symbiosis of leguminous plants with the bacteria collectively called Rhizobium exemplifies this type of liaison (Madsen et al., 2010; Oldroyd et al., 2011). Nitrogen-fixing nodular symbiosis is not ubiquitous, and although pertinent to most legumes it is known in only a very limited number of non-leguminous plants that all belong to a single phylogenetic clade (Soltis et al., 1995; Doyle, 2011). Uncovering the attributes which shape the ability of plants to host nitrogen-fixing bacteria constitutes a subject of intense study (Charpentier and Oldroyd, 2010; Held et al., 2010; Desbrosses and Stougaard, 2011).
In most legume–Rhizobium systems the host plant permits rhizobia to enter root tissues through a highly orchestrated process involving the development of plant plasma membrane-derived conduits called infection threads (ITs) (Jones et al., 2007; Fournier et al., 2008). This process is initiated in response to a chemical tête-à-tête between the interacting partners which, if successful, culminates in the production of rhizobially derived lipo-chitooligosaccharide signaling molecules known as nodulation factors (NFs) (Lerouge et al., 1990; Bek et al., 2010). The perception of NFs by specific plant receptors (Madsen et al., 2003; Radutoiu et al., 2003; Broghammer et al., 2012) incites, among other things, the formation of ITs, although the physical presence of bacteria is required for this process to take place.
Infection threads are guided inside the host roots (van Brussel et al., 1992; Timmers et al., 1999). They extend transcellularly, through a tip growth-like mechanism, from the epidermis or subepidermal cortex towards a subtending region of dividing cortical cells that have initiated the formation of a nodule primordium (NP) (Brewin, 2004; Fournier et al., 2008). Rhizobia are released from ITs inside a subset of NP cells, where they undergo differentiation to bacteroids in confined organelle-like compartments called symbiosomes (Kereszt et al., 2011). In functional nodules, bacteroids fix atmospheric nitrogen to ammonia (Oldroyd et al., 2009). This is supplied for utilization by the plant at the expense of photosynthetic carbon, which is provided by the host to support bacterial respiration (Oldroyd et al., 2005).
The mechanism by which plants regulate the intracellular uptake of symbiotic bacteria, while only partially understood (Jones et al., 2007; Held et al., 2010; Madsen et al., 2010; Murray, 2011), has been shown to primarily depend on various outputs that are associated with the perception of NF and resultant downstream signaling (Oldroyd et al., 2009; Sieberer et al., 2012; Xie et al., 2012; Liang et al., 2013). A key step in this process is the NF-dependent activation of an ancient root response pathway called the common symbiosis pathway, which also supports endosymbiosis with arbuscular mycorrhizal fungi (Duc et al., 1989; Kistner et al., 2005; Parniske, 2008; Bonfante and Genre, 2010). Recent data have highlighted the significance of NF-dependent calcium influx (Morieri et al., 2013) and changes to the actin cytoskeleton (Yokota et al., 2009; Hossain et al., 2012) in mediating rhizobial infection.
Another important aspect of intracellular accommodation is the relationship between infection and the formation of a NP (Oldroyd and Downie, 2008; Madsen et al., 2010). Although empty nodule structures can be induced on legume roots in the absence of rhizobia (Gleason et al., 2006; Tirichine et al., 2006; Hayashi et al., 2010), the presence of infection has been shown to affect nodule organogenesis (Guan et al., 2013). Conversely, there is evidence that events associated with NP formation participate in directing rhizobial infection. For example, in Lotus japonicus, which develops determinate nodules (Szczyglowski et al., 1998), impairment of the Lhk1 cytokinin receptor gene leads to an initial absence of NP, but roots of the lhk1-1 mutant become hyperinfected by Mesorhizobium loti (Murray et al., 2007). The progression of ITs inside the root cortex is more profuse in lhk1-1 than in wild-type L. japonicus but it is also aberrant. This is reflected by a transient stalling of ITs, such that they loop within the root epidermis or subepidermal cortex, appearing to have lost direction (Murray et al., 2007). In the Medicago truncatula Mtcre1 mutant, which carries a deleterious mutation in the presumed ortholog of Lhk1 root hair ITs are formed but their further progression is hindered such that infection mostly fails to enter the root cortex (Gonzalez-Rizzo et al., 2006; Plet et al., 2011). The more restrictive phenotype of the M. truncatula Mtcre1 mutant reflects perhaps a closer relationship between bacterial infection and NP formation during indeterminate nodule formation. Nevertheless, observations in both L. japonicus and M. truncatula indicate that differentiation of NP is pertinent to developing infections, regardless of the type of nodule formed.
In the same context, diploid NP cells appear to be unsuitable for the accommodation of bacteria (Truchet, 1978; Gonzalez-Sama et al., 2006). Progression of a subset of NP cells through cessation of the cell cycle and the subsequent endocycle-dependent increase in nuclear DNA content and cell volume have been considered as essential for the differentiation of infected nodule cells (Cebolla et al., 1999; Vinardell et al., 2003; Gonzalez-Sama et al., 2006). We describe here the identification of a L. japonicus symbiotic mutant called suner1-1 that exhibits a transient defect in nodule development. We show that the underlying mutation affects a gene encoding a predicted subunit A of a topoisomerase VI (TOPO 6A). Our data demonstrate that in L. japonicus the presence of intact SUNER1 TOP6A is required for normal progression of rhizobial infection and timely differentiation of nitrogen-fixing nodules.
Results
Lotus japonicus sup8-1A mutant line
Among the many symbiotic variants identified through a genetic screen for suppressors of the L. japonicus har1-1 hypernodulation phenotype (Murray et al., 2006), a line called suppressor8-1A (sup8-1A) was selected for further analysis. At 14 days after inoculation (dai) with M. loti, the sup8-1A mutant showed an attenuated nodule development phenotype, which was also reflected by enhanced root elongation in comparison with the parental har1-1 line (Figure1a, b). Mapping experiments positioned the presumed ‘suppressor’ locus to a unique location on L. japonicus chromosome 5, which was subsequently confirmed by the identification of the causative gene. For reasons described below, this gene was named SUNERGOS1 (SUNER1) and the corresponding mutant allele is referred to hereafter as suner1-1.
Figure 1.
The Lotus japonicus sup8-1A (suner1-1 har1-1) mutant.(a)–(d) At 14 days after inoculation (dai) with Mesorhizobium loti, the har1-1 parental line (a) develops numerous nodules, which also limits root and shoot growth. The sup8-1A mutant (b) shows attenuated nodule development and enhanced root elongation. In both the parental har1-1 (c) and sup8-1A (d) lines, cell divisions for nodule primordium (NP) formation and root infection by M. loti (blue color) are initiated; however, these are significantly attenuated in sup8-1A and the infection process is stalled either at the epidermis or within the outer cortex above the subtending NP. The double-headed arrow in panel (d) points to a root hair infection thread (IT), while open arrowheads signify aberrant cortical infection events. Root segments were sectioned longitudinally following histochemical staining at 7 dai with M. loti carrying the hemA:LacZ reporter gene (c, d).(e) Scores of nodulation events at 7, 14 and 21 dai; values represent the mean value ± 95% CI (n = 10). Asterisks denote a statistically significant difference (Student's t-test, P ≤ 0.05).
suner1-1 attenuates symbiotic development
To gain insight into the underlying symbiotic defect in sup8-1A (i.e. the suner1-1 har1-1 double mutant), roots from seedlings inoculated with a M. loti strain carrying the hemA:LacZ reporter gene fusion were analyzed. Root hair infections and subtending cortical cell divisions for NP organogenesis were present in the double mutant. However, unlike in har1-1, where at 7 dai the localized cortical cell divisions led to formation of well-defined NP and colonized nodules (Figures1c), these processes were diminished in suner1-1 har1-1. The most obvious differences were the presence of a more compact NP and a significantly decreased number of nodules (Figure1d, e), suggesting a developmental defect in suner1-1 har1-1. The extent of root colonization by M. loti at 7 dai also appeared to be diminished in the mutant. Infection threads readily formed within root hairs but were misdirected and less ramified within the subtending NP (Figure1d).
During subsequent stages, at 14 and 21 dai, the nodulation phenotype of the mutant increasingly resembled that of the parental line, suggesting that the apparent early symbiotic defect in suner1-1 har1-1 was being overcome (Figure1e).
Taken together, these observations suggested that although epidermal infection and cortical cell divisions are initiated in suner1-1 har1-1, their subsequent progression is temporarily halted as if these two converging symbiotic processes have become incompatible or lost their synergy. Reflective of this fact, the underlying locus has been named SUNERGOS1 (SUNER1), from the Greek word syn-ergos, meaning ‘working together’.
The suner1-1 single mutant phenotype
Like suner1-1 har1-1, the suner1-1 single mutant carrying the wild-type homozygous HAR1 locus initially showed a low nodule number phenotype, even though NP were readily formed. At 7 dai, the average number of nodules was significantly lower in suner1-1 compared with wild-type L. japonicus Gifu of the same age (Figure2a). This difference, however, rapidly diminished, such that at 14 dai the average number of nodules in suner1-1 was approximately 65% of the wild-type number. At 21 dai, the suner1-1 nodulation phenotype was not significantly different from the wild type (Figure2a).
Figure 2.

suner1-1 shows only a transient mutant symbiotic phenotype.(a) Scores of nodule primordia and nodules formed at 7, 14 and 21 days after inoculation (dai) in wild-type Lotus japonicus Gifu and suner 1-1; values represent the mean ± 95% CI (n = 10). Asterisks denote a statistically significant difference (Student's t-test P ≤ 0.05).(b)–(d) Three different stages of wild-type nodule development. (e)–(g) The corresponding stages in suner1-1. Note misdirected infection threads (IT) in suner1-1 (e) (blue and arrowhead).
In wild-type plants, ITs readily penetrate subtending regions of cortical cell divisions (Figure2b) where they ramify within the NP (Figure2c) to form fully colonized nodules (Figure2d). These events were also present in suner1-1 (Figure2e–g) except that, in at least some instances, the progression of the infection appeared to be stalled at the interface between the epidermis and cortex or within the subepidermal cortex above the subtending NP (Figure2e). As in suner1-1 har1-1, misdirected ITs were present in suner1-1, migrating parallel to the longitudinal axis of the root instead of growing inside the subtending NP (see below). This defect was rapidly overcome, such that only a few such aberrant infection events could be found in individual suner1-1 roots and wild-type nodules were readily formed (Figure2f, g). It is worth noting that the average number of epidermal ITs in suner1-1 was not significantly different from that in the wild type (Figure S1).
The suner1-1 mutation is temperature sensitive
Given the subtle and ephemeral symbiotic deficiency in suner1-1, we sought conditions that could enhance the mutant phenotype. One approach taken was to examine whether the suner1-1 mutation is temperature sensitive. Indeed, when grown at 28°C suner1-1 showed an enhanced nodulation defect in comparison with plants grown under the usual 21°C regime (Figure3a). At 7 dai this was reflected by the absence of emerged nodules (i.e. those which bulge out of the root epidermis) (Figure3b). Unlike nodules formed 14 dai at 21°C (Figure3a), most emerged nodules on suner1-1 roots grown at 28°C remained small and could readily be distinguished from those of the wild-type control (Figure3b, c).
Figure 3.
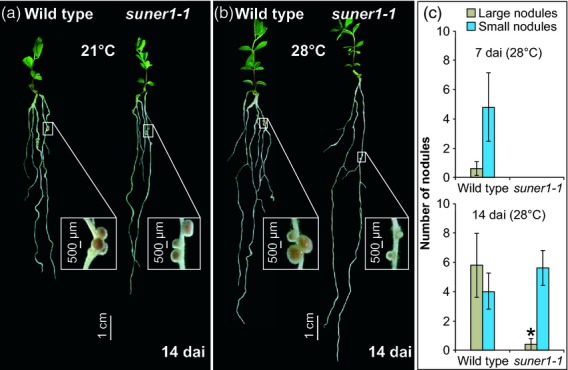
The suner1-1 mutation confers a temperature-sensitive nodulation phenotype.(a) When grown at 21°C the suner1-1 mutant forms mostly wild-type looking nodules.(b) When grown at 28°C the suner1-1 mutant forms mostly small nodules.(c) At 7 and 14 days after inoculation (dai), fully emerged nodules were categorized as either large or small; values in (c) represent the mean ± 95% CI (n = 7). The asterisk indicates a significant difference (Student's t-test, P ≤ 0.05).
Inspection of roots that had been stained for β-galactosidase reporter activity showed that the formation of root hair ITs was unaffected in suner1-1 grown at 28°C. However, as observed at 21°C (Figures2e and 4a), their progression was stalled as they looped within the epidermis or subepidermal cortex above the subtending NP (Figure4b, c). The NP appeared to be even less developed at 28°C in comparison with suner1-1 grown at 21°C (compare Figure4a and b). This suggests that in suner1-1 the increased temperature primarily affects development of the NP and not the behavior of ITs. Importantly, analysis of F2 segregants derived from a cross between suner1-1 and wild-type L. japonicus Gifu (see suner1-1 attenuates symbiotic development) confirmed co-segregation of the homozygous suner1-1 allele with the mutant nodulation phenotype observed at 28°C (Table S1 in Supporting Information). Nevertheless, even under these conditions the suner1-1 mutant phenotype remained ephemeral and mostly wild-type nodules were present at 21 dai.
Figure 4.
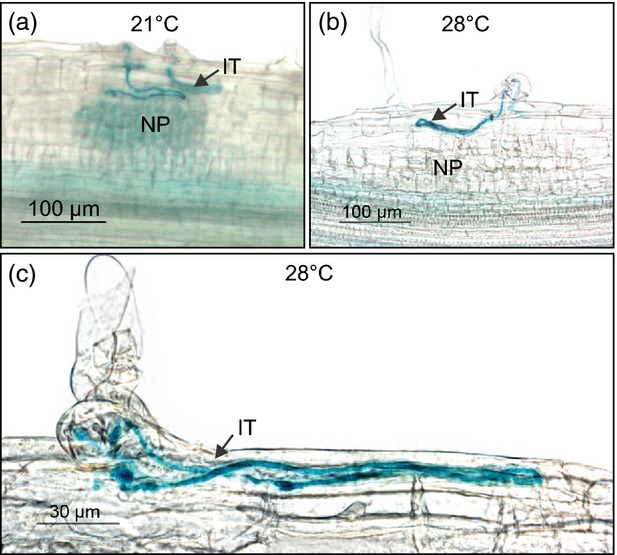
Infection threads are misguided in suner1-1.Seedlings were inoculated with Mesorhizobium loti carrying the hemA:LacZ reporter gene, stained at 7 days after inoculation for β-galactosidase activity and sectioned longitudinally. Note the looping infection threads (IT) within the root epidermis and/or subepidermal cortex above subtending nodule primordia (NP). This phenotype was observed in suner1-1 plants grown at both 21°C (a) and 28°C (b, c).
SUNER1 encodes a predicted subunit A of a topoisomerase VI
A classical linkage analysis positioned the SUNER1 locus on chromosome 5 between flanking markers TM0696 and BM2365 (Figure S2a, b). In parallel, the entire genome of suner1-1 har1-1 was sequenced using next generation sequencing and compared with the wild-type L. japonicus genome sequence. (http://www.kazusa.or.jp/lotus/index.html). A single nucleotide polymorphism (G610 to A) that was well-supported by the sequencing data, with 16× coverage of the relevant region, was located between the two flanking markers. As this polymorphism was associated with a predicted gene model (Figure S2c), this region was considered as a viable candidate for the SUNER1 locus.
The corresponding full-length mRNA was characterized as being 1508 bp long, including 78- and 161-bp 5′ and 3′ untranslated regions (UTRs), respectively (see suner1-1 attenuates symbiotic development). Inspection of the L. japonicus Gene Atlas data (Verdier et al., 2013) showed that this mRNA is present in all L. japonicus tissues tested, including uninoculated roots and nodules (Figure S3).
Alignment of the genomic and mRNA sequences confirmed the structure of the candidate SUNER1 gene as being composed of two exons and one intron (Figure S2c). The SUNER1 transcript contains an open reading frame of 422 amino acids, encoding a predicted protein with a high homology to several known or predicted DNA topoisomerase VI subunit As (TOP6A) from various plants and from Sulfolobus shibatae, an archeobacterium (Figure S4; see also Table S2). SUNER1 shows the highest homology (90% identity and 95% similarity) to a predicted TOP6A from Arachis hypogaea and shares significant homology (85% identity and 93% similarity) with ROOTHAIRLESS2 TOP6A (RHL2) from Arabidopsis thaliana (Table S2). Importantly, all of these proteins, including SUNER1 and the archetypal TOP6A from S. shibatae, share the two-domain structure that is characteristic of TOP6A (Figure S5). They contain the TP6A_N domain, which is thought to be involved in DNA binding based on its sequence similarity to Escherichia coli catabolite activator protein (CAP), and the topoisomerase-primase (TOPPRIM) domain (Aravind et al., 1998; Nichols et al., 1999). The suner1-1 mutation leads to a predicted substitution of the conserved valine204 (V204), which is located close to the C-terminal end of the predicted CAP domain, to methionine (Figure S5).
SUNER1 complements the mutant phenotype
In order to confirm that the correct candidate gene was selected, an in planta complementation experiment was performed. Agrobacterium rhizogenes-mediated transformation of wild-type L. japonicus Gifu shoots produced hairy roots that readily developed large, pink nodules (Figure5a). In contrast, the hairy roots that formed on suner1-1 shoots exaggerated, to some extent, the mutant phenotype forming a decreased number of mostly small nodules (Figure5b). When supplemented with the wild-type copy of the SUNER1 gene, the ability to form large, pink nodules was restored to suner1-1 (Figure5c, d). Taken together the map-based cloning, genome sequencing and complementation analyses showed that the TOP6A-like gene corresponds to the L. japonicus SUNER1 locus.
Figure 5.
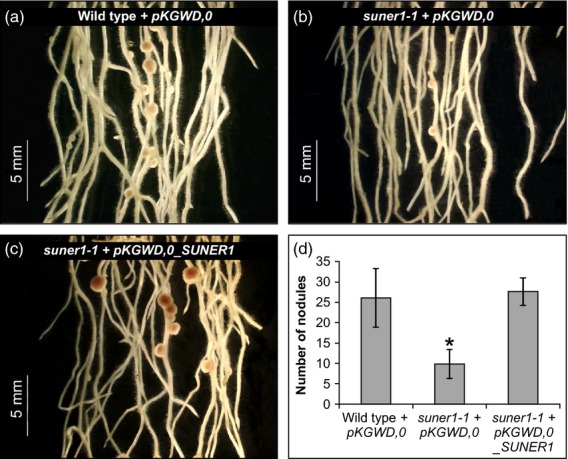
SUNER1 restores wild-type nodule formation to suner1-1. (a)–(c) A genomic fragment containing the entire SUNER1 locus was introduced by Agrobacterium rhizogenes-mediated transformation to generate transgenic hairy roots on non-transgenic shoots. Representative images of transgenic hairy roots that were inoculated with Mesorhizobium loti are shown for the positive control (a, wild-type Gifu + pKGWD,0 empty vector), negative control (b, suner1-1 + pKGWD,0 empty vector) and the complementation experiment (c, suner1-1 + pKGWD,0 containing the SUNER1 gene).(d) Scores of nodules formed on transgenic hairy roots 14 days after inoculation; values represent the mean ± 95% CI (n = 10). The asterisk indicates a significant difference (Student's t-test, P ≤ 0.05).
suner1-1 nodules show decreased levels of 16C and 32C nuclei under elevated temperature
As DNA topoisomerase VI is known to be involved in DNA replication (Yin et al., 2002), we have tested the possibility that SUNER1 TOP6A functions to mediate endoreduplication of nodule cells, presumably beyond the 8C level (Hartung et al., 2002; Sugimoto-Shirasu et al., 2002).
At 14 dai, when grown at 21°C, suner1-1 nodules resemble those of wild-type L. japonicus, but this was the earliest time point when a sufficient amount of nodule tissue could be harvested from unprocessed (i.e. without clearing) L. japonicus roots in order to perform flow cytometry experiments. In L. japonicus wild-type and suner1-1 leaves, approximately 90% of nuclei were at the 2C (diploid) level, with the maximum ploidy at 4C (Figure S6). By contrast, 4C nuclei were the most abundant (about 60%) in uninoculated roots of both genotypes, with the highest ploidy being at 8C. Similar to uninoculated roots, 4C nuclei represented the most abundant category in 14-day-old nodules; however, in both the wild type and suner1-1 the highest ploidy reached the 32C level. In three independent biological replicates, the 2C and 32C nuclei categories appeared somewhat underrepresented in suner1-1 nodules, although the overall peak patterns in leaves and roots were similar between wild type and suner1-1 (Figure S6).
Because the suner1-1 phenotype was more distinct at 14 dai when plants were grown at 28°C (Figure3), the ploidy levels of nodules were re-evaluated under these conditions (Figure6). The distribution of categories of nuclei in wild-type nodules was virtually unchanged from that determined for wild-type plants grown at 21°C (Figures6 and S6). In contrast, suner1-1 nodules showed a shift toward lower ploidy levels, with the presence of only a very small peak for 16C and a total absence of 32C nuclei (Figure6).
Figure 6.
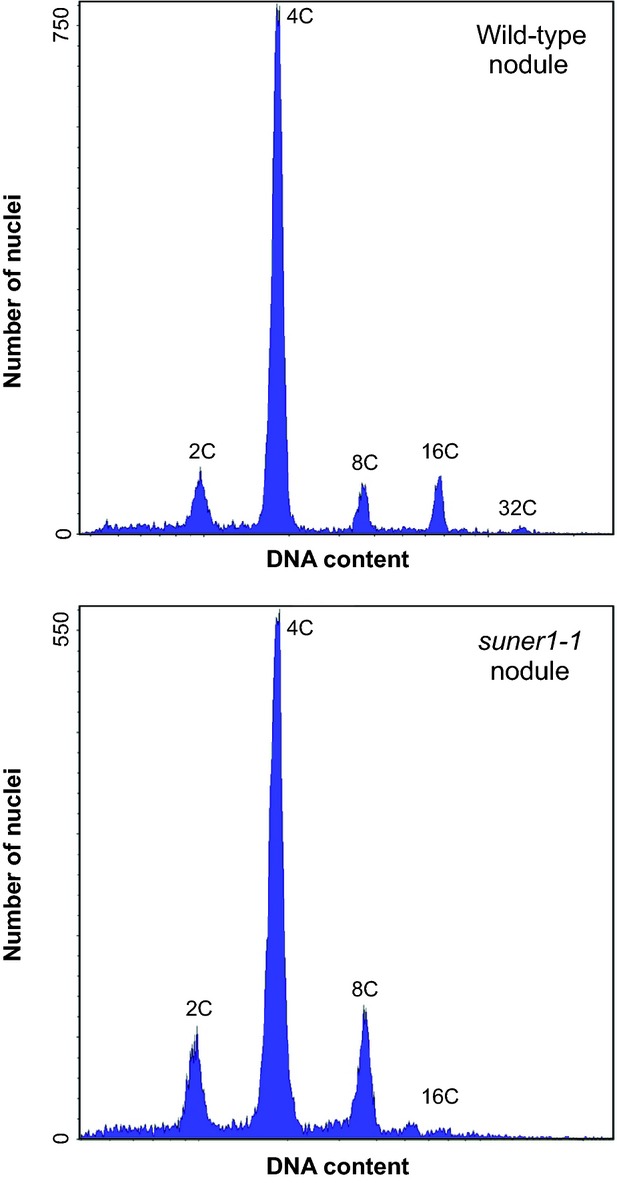
The effect of suner1-1 on the nodule cell ploidy at 28°C.Representative flow cytometric histograms of wild-type (top panel) and suner1-1 (lower panel) nodules are shown. In total, 37 304 and 37 184 nuclei were measured in wild-type and suner1-1 nodules, respectively, in three biological replicates. Note that nodules were collected from roots at 14 days after inoculation.
Discussion
Here, we describe the identification and characterization of a L. japonicus symbiotic mutant that carries the suner1-1 allele. The presence of this allele imposes a weak symbiotic defect, as reflected in a transitory incompatibility between the developing NP and approaching bacterial infection. We show that the L. japonicus SUNER1 locus encodes a predicted subunit A of the TOP6 enzyme, suggesting that the presence of the intact TOP6A is necessary for normal progression of bacterial infection and timely differentiation of nodules.
Suner1 TOP6A
Unlike other eukaryotes, plants contain three different TOP6A homologs (SPO11-1, SPO11-2 and SPO11-3/RHL2) and a single homolog of the B subunit (AtTOP6B) (Hartung and Puchta, 2001). Arabidopsis AtSPO11-3/RHL2 and AtTOP6B interact, forming a functional A2B2 TOP6 enzyme (Sugimoto-Shirasu et al., 2002). However, additional interacting proteins, including ROOT HAIRLESS 1 (RHL1; Sugimoto-Shirasu et al., 2005), MIDGET is the nuclear protein (Kirik et al., 2007), and BIN4, a plant-specific DNA-binding protein (Breuer et al., 2007), were shown to be essential components of an active TOP6 complex.
A great deal of study has been directed towards unraveling the role of TOP6 in plants. In contrast to AtSPO11-1 and AtSPO11-2, which like Saccharomyces cerevisiae SPO11 are involved in meiosis (Keeney et al., 1997; Stacey et al., 2006), AtSPO11-3/RHL2 has an essential function during somatic development (Yin et al., 2002), including cell proliferation, endoreduplication, chromatin remodeling and transcriptional regulation (Hartung et al., 2002; Simková et al., 2012). Typically, deleterious mutations in Arabidopsis AtSPO11-3/RHL2, RHL1, MIDGET, BIN4 or AtTOP6B result in dwarf phenotypes, but they may also cause plant death within a few weeks after germination (Hartung et al., 2002). These effects have been associated primarily with a deficiency in cell proliferation and reduced endoreduplication (Hartung et al., 2002; Sugimoto-Shirasu et al., 2002, 2005; Breuer et al., 2007; Kirik et al., 2007). The functional characterization of suner1-1 highlights the relevance of TOP6A during differentiation of nodules in L. japonicus.
During NP formation, differentiation follows the initial cell divisions and includes endoreduplication of a subset of participating cortical cells (Foucher & Kondorosi, 2000; Kondorosi et al., 2005). The latter process was shown to be essential in establishing functional symbiosis. Downregulation of the anaphase-promoting complex activator CCS52A, which is involved in the transition from mitosis to the endoreduplication cycle, drastically restricts nodule development in M. truncatula (Vinardell et al., 2003). This is reflected by lower cell ploidy, a decreased size of nodules and nodule cells, defects in infection and the inability to establish and/or maintain symbiotic cells, which eventually leads to the death of both bacterial and plant cells (Vinardell et al., 2003). Several of these defects, including the initial formation of small NP and nodules, aberrant infections and lower ploidy levels of nodule cells under elevated temperature, were observed in suner1-1, consistent with the possible role of SUNER1 TOP6A in the differentiation of L. japonicus nodules. This notion is further supported by the recent characterization of the L. japonicus vagrant infection thread 1 (vag1) mutant, which exhibits a strong defect in nodule formation and misdirected ITs. This mutant carries a deleterious mutation in a predicted ortholog of Arabidopsis RHL1, which encodes a component of the TOP6 complex (Dr Takuya Suzaki; National Institute for Basic Biology, Japan, personal communication).
suner1-1 is a weak allele
Unlike downregulation of CCS52A, the suner1-1 mutation does not lead to early senescence of nodule cells. Furthermore, the suner1-1 mutation does not prevent the initial cell divisions for NP formation nor does it have any apparent pleiotropic effect, such as dwarfism.
At the protein level, the suner1-1 mutation is predicted to change V204 to methionine. This substitution is located between the two predicted domains of the protein. Our data, however, point to the functional significance of V204, perhaps consistent with its overall conservation in different species (see Figure S5). Nonetheless, it is likely that the mutant SUNER1-1 TOP6A protein remains active, although not to the same extent as in the wild type. This could explain the weak and ephemeral mutant nodulation phenotype of suner1-1. The same might account for the lack of an apparent pleiotropic effect of the suner1-1 mutation, although an alternative explanation is also possible.
In plants such as A. thaliana, M. truncatula or Lupinus albus different tissues and organs contain highly endoreduplicated nuclei (Kondorosi et al., 2000; Gonzalez-Sama et al., 2006). A survey of various mature L. japonicus tissues, on the other hand, showed that only roots and nodules contain highly polyploid nuclei (Gonzalez-Sama et al., 2006). The results of flow cytometric analyses of L. japonicus leaves, roots and nodules performed during the course of this work (see Figure S6) are consistent with these earlier observations.
If the presence of highly endoreduplicated (i.e. 16C and 32C) nuclei is uniquely or predominantly a feature of nodules in mature L. japonicus plants, this could explain why the effect of the suner1-1 mutation was only observable in this particular tissue. Although TOP6 is involved in the cell cycle and endoreduplication, other DNA topoisomerases can perform these functions (Sugimoto-Shirasu et al., 2005). However, TOP6 appears to be essential for a cell to reach a ploidy level beyond 8C (Sugimoto-Shirasu et al., 2005). Consistent with this prediction, mutations that impair various components of the TOP6 complex lead to the absence of nuclei with higher ploidy (Hartung et al., 2002; Sugimoto-Shirasu et al., 2002, 2005; Yin et al., 2002).
It remains unclear why the suner1-1 mutation lowers the proportion of 2C nuclei, in addition to 32C nuclei, in plants grown at 21°C. Nevertheless, the finding that at an elevated temperature of 28°C the suner1-1 mutation preferentially limits the frequency of 16C and 32C nuclei in nodules might be indicative of the functional relevance of SUNER1 TOP6A during the endoreduplication-dependent differentiation of nodule cells. However, we cannot entirely rule out direct involvement of TOP6A in mediating bacterial infection, although we consider this to be less likely. Root hair ITs form abundantly in suner1-1. Moreover, defects in NP formation have been reported to be associated with the presence of misguided ITs (Murray et al., 2007) and the L. japonicus vag1 mutant displays a strong deficiency in NP formation (see above). It is more likely that the primary defect in suner1-1 is a transitory impairment in the differentiation of NP cells and/or subepidermal cortical cells, which normally act to facilitate the passage of ITs into the deepest region of the root cortex. Whether the observed deficiency in suner1-1 symbiosis is primarily based in the faulty progression through endocycle, as suggested by our flow cytometric data (Figure6), or reflects other cellular functions of TOP6, such as chromatin remodeling and gene silencing, remains to be further established.
Experimental procedures
Plant material, growth conditions and observations of nodulation phenotypes
Lotus japonicus seeds were germinated as described previously (Szczyglowski et al., 1998). Germinated seedlings were transplanted into pots containing sterilized vermiculite:sand (6:1) and were grown at 21 or 28°C under the conditions described in Murray et al. (2006). To visualize the symbiotic bacteria, wild-type and suner1-1 seedlings were inoculated with M. loti strain NZP2235 carrying the hemA::LacZ reporter gene. Root samples were harvested 7, 14 and 21 dai, fixed and histochemically stained for β-galactosidase activity as previously described (Wopereis et al., 2000). In addition to whole root observations, longitudinal and cross-sections of root segments were generated by embedding the stained root in 3% (w/v) agar blocks and sectioning these specimens to a thickness of 30 μm using a Leica VT 1000S vibratome (Leica Microsystems Inc., http://www.leica-microsystems.com/).
A Nikon SMZ1500 (Nikon, http://www.nikon.com/) and a Zeiss Axioskop 2 (Zeiss, http://www.zeiss.com/) were used for microscopic observations. All images were captured with a Nikon DXM1200 digital camera. TIFF format images were created by ACT-1 image software (Nikon).
Nuclear DNA isolation for mapping and next-generation sequencing
Crude genomic DNA preparations were obtained from single leaves using the cetyl-trimethylammonium bromide (CTAB) method (Doyle and Doyle, 1987). To isolate a high molecular weight nuclear DNA for the purpose of next-generation sequencing, approximately 2 g of sup8-1A fresh leaf tissue was pulverized in liquid nitrogen. Powder was suspended in 20 ml of ice-cold 1× HB homogenization buffer (10× HB stock: 0.1 m Trizma Base, 0.8 m KCl, 0.1 m EDTA, 10 mm spermine, 10 mm spermidine) containing 0.5 m sucrose, 0.5% (v/v) Triton X-100 and 0.15% (v/v) β-mercaptoethanol, by gentle swirling on ice for about 10–15 min. The suspension was filtered through two layers of Miracloth and the resulting filtrate was centrifuged at 1800 g in a swinging bucket rotor for 15 min at 4°C. The pellet containing the nuclei was gently washed in 1× HB buffer and centrifuged at 1800 g for 5 min at 4°C. The pellet was resuspended in 500 μl of pre-heated CTAB buffer [2% CTAB (w/v), 100 mm 2-amino-2-(hydroxymethyl)-1,3-propanediol (TRIS)-HCl, pH 8.0, 20 mm EDTA, pH 8.0, and 1.4 m NaCl] and incubated for 30 min at 60°C with gentle shaking at 300 r.p.m. Extraction with 500 μl of chloroform:isoamylalcohol (24:1) was followed by centrifugation at 6000 g for 10 min at 4°C and recovery of the aqueous phase to a new tube. Following RNase treatment at 37°C for 30 min, 0.6 volume of ice-cold isopropanol was added, mixed and incubated for 1 h at −20°C. The DNA was collected by centrifugation at 3500 g for 6 min at 4°C. The resulting pellet was washed with 70% (v/v) ethanol, dried and resuspended in 55 μl of EB buffer (Qiagen, http://www.qiagen.com/).
Map-based cloning of the SUNER1 locus
The rough map position of the SUNER1 locus was initially established using 16 mutants selected from the F2 population obtained by crossing sup8-1A with the polymorphic MG20_har1-1 introgression line (Murray et al., 2006). The same F2 population was utilized for more extensive linkage analysis. In a uniform background of har1-1 (note that both crossing partners carry the homozygous har1-1 mutation) only the suner1-1 allele was expected to segregate. Strict phenotypic selection criteria based on a smaller number of visible nodules and relatively longer roots were applied to select sup8-1A F2 segregants. Approximately 14000 F2 individuals were germinated and 3264 selected mutants were used to analyze recombination events to further delineate the flanking region. Markers used in these analyses were simple sequence repeats (SSRs) obtained from the publicly available L. japonicus genome website (http://www.kazusa.or.jp/lotus/clonelist.html). Polymerase chain reactions (PCRs) were performed using GenScript Taq polymerase (GenScript USA Inc., http://www.genscript.com/). The conditions for amplification of SSR molecular markers were as follows: denaturation at 94°C for 30 sec, annealing at 55°C for 30 sec, extension at 72°C for 30 sec, for a total of 40 cycles, followed by a final extension phase of 72°C for 7 min.
Next-generation sequencing and bioinformatic analyses
Next-generation sequencing of the sup8-1A nuclear DNA was performed in the DNA Technologies Laboratory at the NRC Plant Biotechnology Institute (Saskatoon, Canada) using an Illumina Genome Analyzer IIx sequencer (http://www.illumina.com/). Thirty-seven million reads from two Illumina libraries, a 101-bp paired-end library with 200–300-bp insert size and a 38-bp mate-pair library with a 2.3 kb insert size, were aligned to the L. japonicus MG20 release 2.5 reference genome (http://www.kazusa.or.jp/lotus/) using Bowtie version 0.12.3 (Langmead et al., 2009). The candidate region on chromosome 5 was subsequently annotated for SNPs using SHOREmap (Schneeberger et al., 2009).
Isolation of the suner1-1 single mutant
sup8-1A (i.e. suner1-1 suner1-1/har1-1 har1-1 genotype) was crossed with wild-type L. japonicus Gifu. The resulting F1 plant was self-fertilized to generate a segregating F2 population from which a homozygous suner1-1 single mutant, carrying the wild-type HAR1 locus (i.e. suner1-1 suner1-1/HAR1 HAR1 genotype), was selected. The presence or absence of the har1-1 allele was determined using a cleaved amplified polymorphic marker as previously described (Karas et al., 2005). The SUNER1 locus was genotyped using the method of bidirectional PCR amplification of specific alleles (Bi-PASA) as described in Liu et al. (1997). Two sets of primers, namely the outer SUNER1 Bi-PASA primer pair P and Q and the inner SUNER1 Bi-PASA primer pair A and B were used (see Table S3).
Characterization of the SUNER1 mRNA using 5′ and 3′ rapid amplification of cDNA ends (RACE)
Total RNA was extracted from wild-type roots (14 days after sowing) using the RNeasy Plant Mini kit (Qiagen). First-strand cDNA was generated using the High Capacity cDNA Reverse Transcription Kit with RNase Inhibitor (Applied Biosystems, http://www.appliedbiosystems.com/).
The 5′ and 3′ RACE experiments were performed using the FirstChoice® RLM-RACE kit (Ambion, www.lifetechnologies.com/ca/en/home/brands/ambion.html) following the manufacturer's protocol. The SUNER1 cDNA ends were amplified using the primer pairs as described in Table S3. The amplified PCR fragments were cloned into pGEM®-T Easy vector (Promega, http://www.promega.com/) and sequenced.
In planta complementation experiments
The entire SUNER1 locus (5889 bp), including 3077 and 1409-bp 5′ and 3′ UTRs, respectively, was PCR amplified using the SUNER1 genomic forward and reverse primers (Table S3) and the LjT08O17 genomic contig (http://www.kazusa.or.jp/lotus/release1/predict/cgi-bin/get_seq.cgi?type=fas&db=clone&id=LjT08O17) as the DNA template. The touchdown PCR cycling conditions were as follows: initial denaturation stage of 98°C for 30 sec, a first cycling phase with 5 cycles of 98°C for 10 sec, 60°C for 30 sec, 72°C for 3 min, a second cycling phase with 20 cycles of 98°C for 10 sec, 55°C for 30 sec, 72°C for 3 min, and a final extension period of 72°C for 10 min. The forward primer had been extended by a CACC sequence on the 5′ end to facilitate the directional cloning into the pENTR®/D-TOPO entry vector (Invitrogen, www.lifetechnologies.com/ca/en/home/brands/invitrogen.html). The cloned SUNER1 gene was then recombined from the entry vector through an LR reaction into the pKGWD,0 destination vector (Karimi et al., 2002). The integrity of the complementation construct was confirmed by DNA sequencing and restriction enzyme analyses. Once confirmed, the pKGWD,0–SUNER1 complementation construct was transformed into Agrobacterium rhizogenes strain AR1193. Transformation was conducted as described by Petit et al. (1987) using wild-type and suner1-1 plants. Plants that developed hairy roots at the site of infection were transplanted into sterilized vermiculate soil and grown for 5 days prior to inoculation with wild-type M. loti. Two weeks after inoculation, the nodulation phenotypes of at least 10 independent plants per genotype were evaluated.
Segregation analysis of the temperature-sensitive phenotype
A segregation analysis was performed to confirm that the suner1-1 mutation co-segregates with the mutant phenotype observed at 28°C. A total of 187 F2 individuals segregating the suner1-1 allele were grown at 28°C and categorized based on their nodulation phenotype at 14 dai as either wild type (134 individuals) or suner1-1 (53 individuals). Subsequently, all individuals were subjected to genotyping at the SUNER1 locus. With the exception of six individuals, which had poor nodulation and were initially categorized as suner1-1 but had the wild-type genotype, all remaining, phenotypically selected mutants were confirmed as being homozygous suner1-1 plants while all wild-type individuals were either homozygous dominant (SUNER1 SUNER1) or heterozygous (SUNER1 suner1-1) at the locus. This analysis confirmed co-segregation of the temperature-sensitive phenotype with the suner1-1 mutation while also providing independent confirmation of the recessive monogenic nature of the mutation.
Flow cytometric analyses
Nuclei were extracted from various wild-type and suner1-1 tissues in LB01 buffer (Dolezel et al., 1989). Approximately 100 mg of tissue was submerged in 1 ml of LB01 buffer in a plastic Petri dish and chopped with a razor blade for 30 sec. The homogenate was filtered through CellTrics® 30-μm sample filters (Partec, http://www.partec.com/) yielding approximately 600 μl of filtrate, which was incubated on ice in the dark for 20 min. Following the incubation, the filtered nuclei were analyzed using a BD FACSCalibur Flow Cytometer (eBioscience Inc., http://www.ebioscience.com/). Accession number: The SUNERGOS1 gene sequence can be found in the GenBank under the following accession number: KJ671531.
Acknowledgments
We thank Alex Molnar for his expert help in preparation of the figures. This work was supported by grants from Agriculture and Agri-Food Canada Crop Genomics Initiative and National Science and Engineering Research Council of Canada (NSERC grant no. 3277A01) to KS. MH was supported in part by the NSERC-PGS-D2 fellowship. SUA and JS were supported by the Danish National Research Foundation grant no. DNRF79.
Conflict of interest
The authors have no conflict of interest to declare.
Supporting Information
Additional Supporting Information may be found in the online version of this article.
Figure S1.Number of infection threads in wild-type Lotus japonicus Gifu and suner1-1.
Figure S2. Map-based cloning of the SUNER1 locus.
Figure S3.SUNER1 is expressed in various Lotus japonicus tissues.
Figure S4. SUNER1 shares homology with known TOP6A proteins from different species.
Figure S5. Amino acid sequence alignment.
Figure S6. Ploidy levels in wild-type Lotus japonicus Gifu and suner1-1.
Table S1.Similarities and identities between TOP6A proteins from different organisms.
Table S2. Genotypes of 187 F2 individuals segregating the suner1-1 mutation at 28°C.
Table S3. List of primers.
References
- Aravind L, Leipe DD, Koonin EV. Toprim-a conserved catalytic domain in type IA and II topoisomerases, DnaG-type primases, OLD family nucleases and RecR proteins. Nucleic Acids Res. 1998;26:4205–4213. doi: 10.1093/nar/26.18.4205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bek AS, Sauer J, Thygesen MB, Duus JO, Petersen BO, Thirup S, James E, Jensen KJ, Stougaard J, Radutoiu S. Improved characterization of Nod factors and genetically based variation in LysM receptor domains identify amino acids expendable for Nod factor recognition in Lotus spp. Mol. Plant Microbe Interact. 2010;23:58–66. doi: 10.1094/MPMI-23-1-0058. [DOI] [PubMed] [Google Scholar]
- Bonfante P, Genre A. Mechanisms underlying beneficial plant-fungus interactions in mycorrhizal symbiosis. Nat. Comm. 2010;48:1–11. doi: 10.1038/ncomms1046. [DOI] [PubMed] [Google Scholar]
- Breuer C, Stacey NJ, West CE, Zhao Y, Chory J, Tsukaya H, Azumi Y, Maxwell A, Roberts K, Sugimoto-Shirasua K. BIN4, a novel component of the plant DNA topoisomerase VI complex, is required for endoreduplication in Arabidopsis. Plant Cell. 2007;19:3655–3668. doi: 10.1105/tpc.107.054833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brewin NJ. Plant cell wall remodelling in the Rhizobium-Legume symbiosis. Crit. Rev. Plant Sci. 2004;23:293–316. [Google Scholar]
- Broghammer A, Krusell L, Blaise M, et al. Legume receptors perceive the rhizobial lipochitin oligosaccharide signal molecules by direct binding. Proc. Natl Acad. Sci. USA. 2012;109:13859–13864. doi: 10.1073/pnas.1205171109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Brussel AAN, Bakhuizen R, van Spronsen PC, Spaink HP, Tak T, Lugtenberg BJJ, Kijne JW. Induction of preinfection thread structures in the leguminous host plant by mitogenic lipo-oligosaccharides of Rhizobium. Science. 1992;257:70–72. doi: 10.1126/science.257.5066.70. [DOI] [PubMed] [Google Scholar]
- Cebolla A, Vinardell JM, Kiss E, Oláh B, Roudier F, Kondorosi A, Kondorosi E. The mitotic inhibitor ccs52 is required for endoreduplication and ploidy-dependent cell enlargement in plants. EMBO J. 1999;18:4476–4484. doi: 10.1093/emboj/18.16.4476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charpentier M, Oldroyd G. How close are we to nitrogen-fixing cereals? Curr. Opin. Plant Biol. 2010;13:556–564. doi: 10.1016/j.pbi.2010.08.003. [DOI] [PubMed] [Google Scholar]
- Desbrosses GJ, Stougaard J. Root nodulation, a paradigm for how plant-microbe symbiosis influences host developmental pathways. Cell Host Microbe. 2011;10:348–358. doi: 10.1016/j.chom.2011.09.005. [DOI] [PubMed] [Google Scholar]
- Dolezel J, Binarova P, Lucretti S. Analysis of nuclear DNA content in plant cells by flow cytometry. Biol. Planta. 1989;31:113–120. [Google Scholar]
- Doyle JJ. Phylogenic perspectives on the origin of nodulation. Mol. Plant Microbe Interact. 2011;24:1289–1295. doi: 10.1094/MPMI-05-11-0114. [DOI] [PubMed] [Google Scholar]
- Doyle JJ, Doyle JL. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem. Bull. 1987;19:11–15. [Google Scholar]
- Duc G, Trouvelot A, Gianinazzi-Pearson V, Gianinazzi S. First report of non-mycorrhizal plant mutants (Myc−) obtained in pea (Pisum sativum L.) and fababean (Vicia faba L.) Plant Sci. 1989;60:215–222. [Google Scholar]
- Foucher F, Kondorosi E. Cell cycle regulation in the course of nodule organogenesis in Medicago. Plant Mol. Biol. 2000;43:773–786. doi: 10.1023/a:1006405029600. [DOI] [PubMed] [Google Scholar]
- Fournier J, Timmers ACJ, Sieberer BJ, Jauneau A, Chabaud M, Barker DG. Mechanism of infection thread elongation in root hairs of Medicago truncatula and dynamic interplay with associated rhizobial colonization. Plant Physiol. 2008;148:1985–1995. doi: 10.1104/pp.108.125674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gleason C, Chaudhuri S, Yang T, Munoz A, Poovaiah BW, Oldroyd GED. Nodulation independent of rhizobia induced by a calcium-activated kinase lacking autoinhibition. Nature. 2006;441:1149–1152. doi: 10.1038/nature04812. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Rizzo S, Crespi M, Frugier F. The Medicago truncatula CRE1 cytokinin receptor regulates lateral root development and early symbiotic interaction with Sinorhizobium meliloti. Plant Cell. 2006;18:2680–2693. doi: 10.1105/tpc.106.043778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Sama A, de la Pena TC, Kevei Z, Mergaert P, Lucas MM, de Felipe MR, Kondorosi E, Pueyo JJ. Nuclear DNA endoreduplication and expression of the mitotic inhibitor Ccs52 associated to determinate and lupinoid nodule organogenesis. Mol. Plant Microbe Interact. 2006;19:173–180. doi: 10.1094/MPMI-19-0173. [DOI] [PubMed] [Google Scholar]
- Guan D, Stacey N, Liu C, et al. Rhizobial infection is associated with the development of peripheral vasculature in nodules of Medicago truncatula. Plant Physiol. 2013;162:107–115. doi: 10.1104/pp.113.215111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartung F, Puchta H. Molecular characterization of homologues of both subunits A (SPO11) and B of the archaebacterial topoisomerase 6 in plants. Gene. 2001;271:81–86. doi: 10.1016/s0378-1119(01)00496-6. [DOI] [PubMed] [Google Scholar]
- Hartung F, Angelis KJ, Meister A, Schubert I, Melzer M, Puchta H. An archaebacterial topoisomerase homolog not present in other eukaryotes is indispensable for cell proliferation of plants. Curr. Biol. 2002;12:1787–1791. doi: 10.1016/s0960-9822(02)01218-6. [DOI] [PubMed] [Google Scholar]
- Hayashi T, Banba M, Shimoda Y, Kouchi H, Hayashi M, Imaizumi-Anraku H. A dominant function of CCaMK in intracellular accommodation of bacterial and fungal endosymbionts. Plant J. 2010;63:141–154. doi: 10.1111/j.1365-313X.2010.04228.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Held M, Hossain MdS, Yokota K, Bonfante P, Stougaard J, Szczyglowski K. Common and not so common symbiotic entry. Trends Plant Sci. 2010;15:540–545. doi: 10.1016/j.tplants.2010.08.001. [DOI] [PubMed] [Google Scholar]
- Hossain MdS, Liao J, James EK, Sato S, Tabata S, Jurkiewicz A, Madsen LH, Stougaard J, Ross L, Szczyglowski K. Lotus japonicus ARPC1 is required for rhizobial infection. Plant Physiol. 2012;160:917–928. doi: 10.1104/pp.112.202572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones KM, Kobayashi H, Davies BW, Taga ME, Walker GC. How rhizobial symbionts invade plants: the SinorhizobiumMedicago model. Nature Rev. Microbiol. 2007;5:619–633. doi: 10.1038/nrmicro1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karas B, Murray J, Gorzelak M, Smith A, Sato S, Tabata S, Szczyglowski K. Invasion of Lotus japonicus root hairless 1 by Mesorhizobium loti involves the nodulation factor-dependent induction of root hairs. Plant Physiol. 2005;137:1331–1344. doi: 10.1104/pp.104.057513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi M, Inze D, Depicker A. GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 2002;7:193–195. doi: 10.1016/s1360-1385(02)02251-3. [DOI] [PubMed] [Google Scholar]
- Keeney S, Giroux CN, Kleckner N. Meiosis-specific DNA double-strand breaks are catalyzed by Spo11, a member of a widely conserved protein family. Cell. 1997;88:375–384. doi: 10.1016/s0092-8674(00)81876-0. [DOI] [PubMed] [Google Scholar]
- Kereszt A, Mergaert P, Kondorosi E. Bacteroid development in legume nodules: evolution of mutual benefit or of sacrificial victims. Mol. Plant Microbe Interact. 2011;24:1300–1309. doi: 10.1094/MPMI-06-11-0152. [DOI] [PubMed] [Google Scholar]
- Kirik V, Schrader A, Uhrig JF, Hulskamp M. MIDGET unravels functions of the Arabidopsis topoisomerase VI complex in DNA endoreduplication, chromatin condensation, and transcriptional silencing. Plant Cell. 2007;19:3100–3110. doi: 10.1105/tpc.107.054361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kistner C, Winzer T, Pitzschke A, et al. Seven Lotus japonicus genes required for transcriptional reprogramming of the root during fungal and bacterial symbiosis. Plant Cell. 2005;17:2217–2229. doi: 10.1105/tpc.105.032714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondorosi E, Roudier F, Gendreau E. Plant cell-size control: growing by ploidy? Curr. Opin. Plant Biol. 2000;3:488–492. doi: 10.1016/s1369-5266(00)00118-7. [DOI] [PubMed] [Google Scholar]
- Kondorosi A, Vinardell JM, Uchiumi T, Mergaert P, Kondorosi E. Cell cycle and symbiosis. Curr. Plant Sci. Biotech. Agric. 2005;41:147–151. [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerouge P, Roche P, Faucher C, Maillet F, Truchet G, Promé JC, Dénarié J. Symbiotic host-specificity of Rhizobium meliloti is determined by a sulphated and acylated glucosamine oligosaccharide signal. Nature. 1990;344:781–784. doi: 10.1038/344781a0. [DOI] [PubMed] [Google Scholar]
- Liang Y, Cao Y, Tanaka K, Thibivilliers S, Wan J, Choi J, Kang C, Qiu J, Stacey G. Nonlegumes respond to rhizobial Nod factors by suppressing the innate immune response. Science. 2013;341:1384–1387. doi: 10.1126/science.1242736. [DOI] [PubMed] [Google Scholar]
- Liu Q, Thorland EC, Heit JA, Sommer SS. Overlapping PCR for bidirectional PCR amplification of specific alleles: a rapid one-tube method for simultaneously differentiating homozygotes and heterozygotes. Genome Res. 1997;7:389–398. doi: 10.1101/gr.7.4.389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madsen EB, Madsen LH, Radutoiu S, et al. A receptor kinase gene of the LysM type is involved in legume perception of rhizobial signals. Nature. 2003;425:637–640. doi: 10.1038/nature02045. [DOI] [PubMed] [Google Scholar]
- Madsen LH, Tirichine L, Jurkiewicz A, Sullivan JT, Heckmann AB, Bek AS, Ronson CW, James EK, Stougaard J. The molecular network governing nodule organogenesis and infection in the model legume Lotus japonicus. Nat. Commun. 2010;10:1–12. doi: 10.1038/ncomms1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morieri G, Martinez EA, Jarynowski A, Driguez H, Morris R, Oldroyd GE, Downie JA. Host-specific Nod-factors associated with Medicago truncatula nodule infection differentially induce calcium influx and calcium spiking in root hairs. New Phytol. 2013;200:656–662. doi: 10.1111/nph.12475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray JD. Invasion by invitation: rhizobial infection in legumes. Mol. Plant Microbe Interact. 2011;24:631–639. doi: 10.1094/MPMI-08-10-0181. [DOI] [PubMed] [Google Scholar]
- Murray J, Karas B, Ross L, et al. Genetic suppressors of the Lotus japonicus har1-1 hypernodulation phenotype. Mol. Plant Microbe Interact. 2006;19:1082–1091. doi: 10.1094/MPMI-19-1082. [DOI] [PubMed] [Google Scholar]
- Murray J, Karas B, Sato S, Tabata S, Amyot L, Szczyglowski K. A cytokinin perception mutant colonized by Rhizobium in the absence of nodule organogenesis. Science. 2007;315:101–104. doi: 10.1126/science.1132514. [DOI] [PubMed] [Google Scholar]
- Nichols MD, DeAngelis K, Keck JL, Berger JM. Structure and function of an archaeal topoisomerase VI subunit with homology to the meiotic recombination factor Spo11. EMBO J. 1999;18:6177–6188. doi: 10.1093/emboj/18.21.6177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldroyd GED, Downie JA. Coordinating nodule morphogenesis with rhizobial infection in legumes. Annu. Rev. Plant Biol. 2008;59:519–546. doi: 10.1146/annurev.arplant.59.032607.092839. [DOI] [PubMed] [Google Scholar]
- Oldroyd G, Harrison M, Udvardi M. Peace talks and trade deals. Keys to long term harmony in legume-microbe symbioses. Plant Physiol. 2005;137:1205–1210. doi: 10.1104/pp.104.057661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldroyd GED, Harrison MJ, Paszkowki U. Reprogramming plant cell for endosymbiosis. Science. 2009;324:753–754. doi: 10.1126/science.1171644. [DOI] [PubMed] [Google Scholar]
- Oldroyd GE, Murray JD, Poole PS, Downie JA. The rules of engagement in the legume-rhizobial symbiosis. Ann. Rev. Genet. 2011;45:119–144. doi: 10.1146/annurev-genet-110410-132549. [DOI] [PubMed] [Google Scholar]
- Parniske M. Arbuscular mycorrhiza: the mother of plant root endosymbioses. Nature. 2008;6:763–775. doi: 10.1038/nrmicro1987. [DOI] [PubMed] [Google Scholar]
- Petit A, Stougaard J, Kühle A, Marcker KA, Tempé J. Transformation and regeneration of the legume Lotus corniculatus: a system for molecular studies of nitrogen fixation. Mol. Gen. Genet. 1987;207:245–250. [Google Scholar]
- Plet J, Wasson A, Ariel F, Le Signor C, Baker D, Mathesius U, Crespi M, Frugier F. MtCRE1-dependent cytokinin signaling integrates bacterial and plant cues to coordinate symbiotic nodule organogenesis in Medicago truncatula. Plant J. 2011;65:622–633. doi: 10.1111/j.1365-313X.2010.04447.x. [DOI] [PubMed] [Google Scholar]
- Radutoiu S, Madsen LH, Madsen EB, et al. Plant recognition of symbiotic bacteria requires two LysM receptor-like kinases. Nature. 2003;425:585–592. doi: 10.1038/nature02039. [DOI] [PubMed] [Google Scholar]
- Schneeberger K, Ossowski S, Lanz C, Juul T, Petersen AH, Nielsen KL, Jørgensen JE, Weigel D, Andersen SU. SHOREmap: simultaneous mapping and mutation identification by deep sequencing. Nat. Methods. 2009;6:550–551. doi: 10.1038/nmeth0809-550. [DOI] [PubMed] [Google Scholar]
- Sieberer BJ, Chabaud M, Fournier J, Timmers ACJ, Barker DG. A switch in Ca2+ spiking is concomitant with endosymbiotic microbe entry into cortical root cells of Medicago truncatula. Plant J. 2012;69:822–830. doi: 10.1111/j.1365-313X.2011.04834.x. [DOI] [PubMed] [Google Scholar]
- Simková K, Moreau F, Pawlak P, Vriet C, Baruah A, Alexandre C, Hennig L, Apel K, Laloi C. Integration of stress-related and reactive oxygen species-mediated signals by Topoisomerase VI in Arabidopsis thaliana. Proc. Natl Acad. Sci. USA. 2012;109:16360–16365. doi: 10.1073/pnas.1202041109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soltis DE, Soltis PS, Morgan DR, Swensen SM, Mullin BC, Dowd JM, Martin PG. Chloroplast gene sequence data suggest a single origin of the predisposition for symbiotic nitrogen-fixation in angiosperms. Proc. Natl Acad. Sci. USA. 1995;92:2647–2651. doi: 10.1073/pnas.92.7.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stacey NJ, Kuromori T, Azumi Y, Roberts G, Breuer C, Wada T, Maxwell A, Roberts K, Sugimoto-Shirasu K. Arabidopsis SPO11-2 functions with SPO11-1 in meiotic recombination. Plant J. 2006;48:206–216. doi: 10.1111/j.1365-313X.2006.02867.x. [DOI] [PubMed] [Google Scholar]
- Sugimoto-Shirasu K, Stacey NJ, Corsar J, Roberts K, McCann MC. DNA topoisomerase VI is essential for endoreduplication in Arabidopsis. Curr. Biol. 2002;12:1782–1786. doi: 10.1016/s0960-9822(02)01198-3. [DOI] [PubMed] [Google Scholar]
- Sugimoto-Shirasu K, Roberts GR, Stacey NJ, McCann MC, Maxwell A, Roberts K. RHL1 is an essential component of the plant DNA topoisomerase VI complex and is required for ploidy-dependent cell growth. Proc. Natl Acad. Sci. USA. 2005;102:18736–18741. doi: 10.1073/pnas.0505883102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szczyglowski K, Shaw SR, Wopereis J, Hamburger D, Copeland S, Dazzo FB, de Bruijn FJ. Nodule organogenesis and symbiotic mutants of the model legume Lotus japonicus. Mol. Plant-Microbe Interact. 1998;11:684–697. [Google Scholar]
- Timmers ACJ, Auriac MC, Truchet G. Refined analysis of early symbiotic steps of the Rhizobium-Medicago interaction in relationship with microtubular cytoskeleton rearrangements. Development. 1999;126:3617–3628. doi: 10.1242/dev.126.16.3617. [DOI] [PubMed] [Google Scholar]
- Tirichine L, Imaizumi-Anraku H, Yoshida S, et al. Deregulation of a Ca2+/calmodulin-dependent kinase leads to spontaneous nodule development. Nature. 2006;441:1153–1156. doi: 10.1038/nature04862. [DOI] [PubMed] [Google Scholar]
- Truchet G. Sur l’état diploide des cellules du méritème des nodules radiculaires des légumineuses. Ann. Sci. Nat. Bot. Paris. 1978;19:3–38. [Google Scholar]
- Verdier J, Torres-Jerez I, Wang M, Andriankaja A, Allen SN, He J, Tang Y, Murray JD, Udvardi MK. Establishment of the Lotus japonicus Gene Expression Atlas (LjGEA) and its use to explore legume seed maturation. Plant J. 2013;74:351–362. doi: 10.1111/tpj.12119. [DOI] [PubMed] [Google Scholar]
- Vinardell JM, Fedorova E, Cebolla A, et al. Endoreduplication mediated by the anaphase-promoting complex activator CCS52A is required for symbiotic cell differentiation in Medicago truncatula nodules. Plant Cell. 2003;15:2093–2105. doi: 10.1105/tpc.014373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wopereis J, Pajuelo E, Dazzo FB, Jiang QY, Gresshoff PM, de Bruijn FJ, Stougaard J, Szczyglowski K. Short root mutant of Lotus japonicus with a dramatically altered symbiotic phenotype. Plant J. 2000;23:97–114. doi: 10.1046/j.1365-313x.2000.00799.x. [DOI] [PubMed] [Google Scholar]
- Xie F, Murray JD, Kim J, Heckmann AB, Edwards A, Oldroyd GE, Downie JA. Legume pectate lyase required for root infection by rhizobia. Proc. Natl Acad. Sci. USA. 2012;109:633–638. doi: 10.1073/pnas.1113992109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin YH, Cheong H, Friedrichsen D, Zhao YD, Hu JP, Mora-Garcia S, Chory J. A crucial role for the putative Arabidopsis topoisomerase VI in plant growth and development. Proc. Natl Acad. Sci. USA. 2002;99:10191–10196. doi: 10.1073/pnas.152337599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokota K, Fukai E, Madsen LH, et al. NAP and PIR-dependent rearrangement of actin cytoskeleton mediates invasion of Lotus japonicus roots by Mesorhizobium loti. Plant Cell. 2009;21:267–284. doi: 10.1105/tpc.108.063693. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1.Number of infection threads in wild-type Lotus japonicus Gifu and suner1-1.
Figure S2. Map-based cloning of the SUNER1 locus.
Figure S3.SUNER1 is expressed in various Lotus japonicus tissues.
Figure S4. SUNER1 shares homology with known TOP6A proteins from different species.
Figure S5. Amino acid sequence alignment.
Figure S6. Ploidy levels in wild-type Lotus japonicus Gifu and suner1-1.
Table S1.Similarities and identities between TOP6A proteins from different organisms.
Table S2. Genotypes of 187 F2 individuals segregating the suner1-1 mutation at 28°C.
Table S3. List of primers.



