Figure 1.
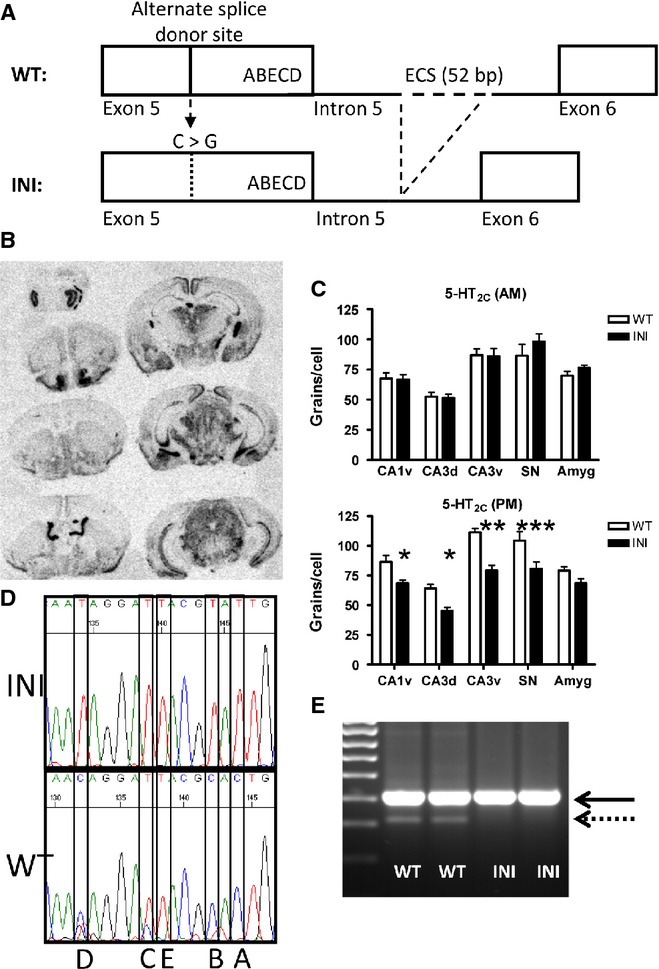
Generation and verification of the INI mouse model. (A) Schematic representation of the targeting strategy used for the INI mice generation. Editing at five sites (A, B, E, C and D) in exon 5 of Htr2c was prevented by deleting the exon complementary sequence (ECS) situated in the adjacent intron, thereby inhibiting the formation of a double-stranded RNA structure and the action of the ADAR enzyme (Adenosine Deaminase Acting on RNA). The alternate splice donor site was mutated to prevent the splicing of the transcript. (B) In situ hybridisation shows that the brain pattern of INI Htr2c RNA expression is normal. (C) Morning and evening levels of Htr2c mRNA were quantified from the in situ hybridisation; the transcript was differentially expressed in the evening only (n = 7–11; *P < 0.05, **P < 0.01, ***P < 0.001). (D) Sequencing traces generated from reverse-transcribed RNA (complementary sequence shown, T and C correspond to an A and G in the Htr2c coding sequence) and showing the absence of editing in the INI animals at the five sites (A, B, E, C and D). (E) Following reverse transcription–polymerase chain reaction of Htr2c transcripts, this gel shows that the full-length receptor variant is expressed (411 bp, solid line) and the truncated splice variant (dotted line) is missing from the INI mouse RNA (see text for details). SN, Substantia Nigra.
