FIGURE 4.
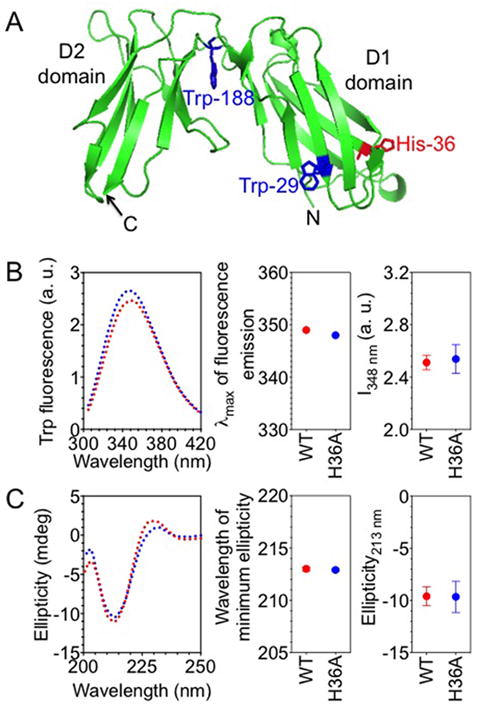
H36A mutation does not perturb the overall structure of KIR2DL1. (A) Ribbon diagram structure of KIR2DL1. Tryptophan residues 29, 188, and His-36 are shown as sticks. Tryptophan residue 207 is not visible in KIR2DL1 structure, probably due to a disordered stem (201–224) structure. The diagram was generated from the PDB file 1NKR using pymol. (B) Intrinsic tryptophan fluorescence of purified, soluble KIR2DL1. The left panel shows the spectra of the wild type (2DL1-WT) (red) and the H36A mutant (2DL1-H36A) (blue) of KIR2DL1 (1-224). Maximum wavelengths of emission and fluorescence intensities at 348 nm (I348 nm) are shown in the middle and the right-most panels, respectively. The error bars represent spread in the values of mean determined from two independent experiments. (C) Far-UV circular dichroism of purified, soluble KIR2DL1. The left panel shows the far-UV circular dichroism spectra of 2DL1-WT (red) and 2DL1-H36A (blue). The positions of minimum ellipticity and the ellipticity values at 213 nm are plotted in the middle and the right-most panels, respectively. The error bars represent spread in the values of mean determined from two independent experiments.
