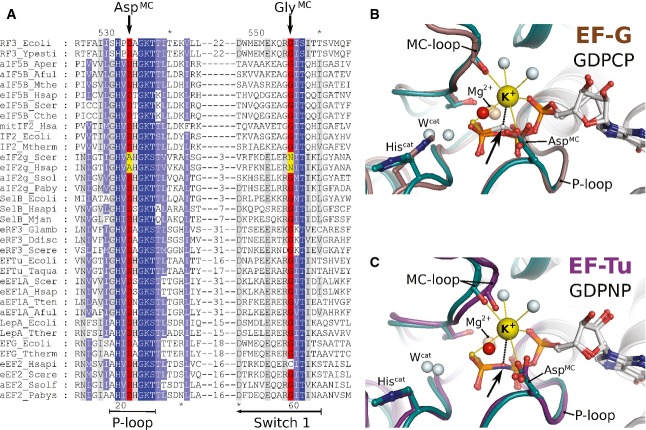
Figure 3. Structural elements required for M+-coordination in eIF5B are universally conserved among trGTPases.
- Excerpt of a multiple sequence alignment of different trGTPases (orthologs of RF3, eIF5B, eIF2γ, SelB, eRF3, EF-Tu, LepA, EF-G) showing P-loop and switch 1. The upper and lower numbering corresponds to C. thermophilum eIF5B (Cthe) and Escherichia coli EF-Tu (Ecoli), respectively. Highly conserved residues are highlighted in dark blue, conserved residues in light blue. AspMC and GlyMC are highlighted in red; residues in eIF2γ that replace AspMC and GlyMC are highlighted in yellow.
- Superposition of eIF5B·GTPγS·K+ (colored as in Fig 1) with ribosome-bound EF-G·GDPCP (brown; PDB: 4JUW). Ribosome-bound EF-G provides all structural elements to bind the M+ ion; however, its coordination is prevented by the CH2 group of GDPCP in lieu of the β-γ-bridging oxygen (arrow). A water molecule (red sphere) is bound next to the M+-binding site instead.
- Similarly, EF-Tu·GDPNP (purple; PDB: 2C78) provides all structural elements to bind the M+ ion; however, its coordination is prevented by the NH group of GDPNP (arrow). A water molecule (red sphere) is bound next to the M+ binding site instead.