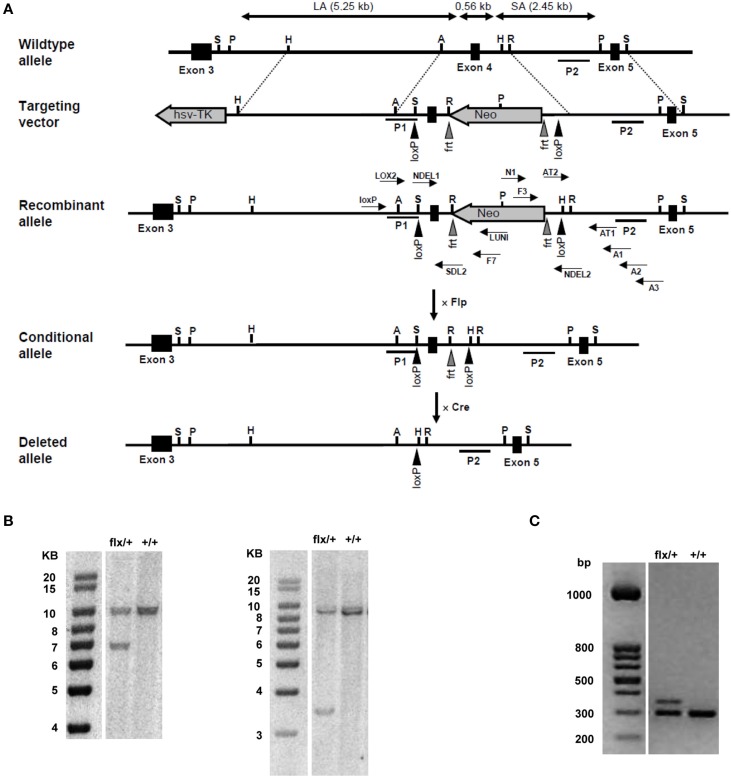
Figure 1.
Generation of NFAT5 conditional null allele. (A) Schematic figures of the wild-type NFAT5 allele, the targeting vector, and the targeted NFAT5neo-flx, NFAT5flx, and NFAT5Δ alleles. Exons are represented by open boxes in which the exon number is indicated. Black triangles denote loxP sites; gray triangles denote frt sites. The 5′ homologous arm of the targeting vector is ~ 5.25 kb, the 3′ homologous arm ~ 2.45 kb. P1 and P2 denote the location of the 5′- and the 3′- external probe used for Southern blot analysis. Arrows indicate the position of the primers used for gene targeting and genotyping. NFAT5neo-flx mice were mated with Flp mice to generate NFAT5flx mice. NFAT5Δ mice were generated by crossing NFAT5flx mice with transgenic Cre mice and subsequent tamoxifen administration to targeted offspring. (B) Southern blot analysis of NFAT5neo-flx/+ genomic DNA. 5′- and 3′- external probes were used to detect the presence of the LA (5.25 kb) and the SA (2.45 kb) of the targeted NFAT5 fragment. (C) Genotypes of mice were identified using genomic DNA prepared from tail biopsies. Primers LOX2 and SDL2 were used to distinguish the targeted allele (392 bp) from the untargeted wild-type allele (334 bp).