Figure 1.
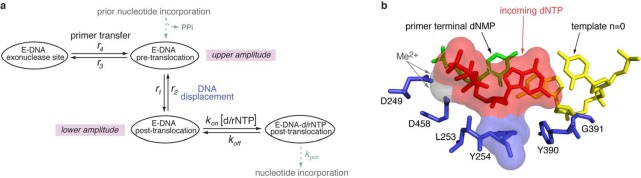
Kinetic and structural determinants of stable dNTP or rNTP incorporation. (a) Model for the kinetic relationships among the steps of translocation, primer strand transfer between the polymerase and exonuclease active sites, and nucleotide triphosphate binding. The kinetic model is fully described by six transition rates: the rates of translocation (r1, r2), the rates of primer stand transfer between the pre-translocation state polymerase site and the exonuclease site (r3, r4), and the rates of dNTP binding to post-translocation state complexes (kon[dNTP] and koff). The mathematical framework based on the model allows us to determine these rates from experimental measurements.37,38,40 (b) View of the polymerase active site in the Φ29 DNAP–DNA–dNTP, post-translocation state ternary complex from the crystal structure model in PDBID 2PYJ. The structure is from ref (28) and was determined using the D12A/D66A mutant of Φ29 DNAP. Protein residues are blue, the template strand (the n = 0 and n = −1 template residues) are yellow, the primer terminal residue is green, and the incoming dNTP is shown in red. The two active site Me2+ ions are rendered as gray spheres. The solvent accessible surfaces of the incoming dNTP and residue Y254, in red and blue, respectively, are shown to highlight the stacking of the deoxyribose sugar of the incoming dNTP on the phenyl ring of residue Y254. Surfaces were rendered in PyMol using a solvent radius of 1.4 Å.
