Figure 4.
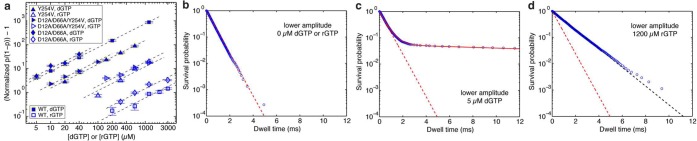
Complementary dNTP or rNTP binding to wild-type Φ29 DNAP and mutants. (a) The normalized p/(1 – p) – 1 is shown in a log–log plot as a function of the concentration of dGTP (filled symbols) or of rGTP (open symbols), for complexes formed between DNA1-H_H and wild-type Φ29 DNAP or mutants. Enzyme identities are indicated by symbol shapes: the wild-type Φ29 DNAP (□), the Y254V mutant (△), the D12A/D66A mutant (◇), or the D12A/D66A/Y254V mutant (▶). The normalized p/(1 – p) is defined as the value of p/(1 – p) in the presence of a given concentration of dNTP or rNTP, divided by the value of p/(1 – p) for the same Φ29 DNAP–DNA complex at 0 μM dNTP or rNTP36where p is the probability of post-translocation state occupancy. Complexes were captured at 180 mV. Error bars show the standard errors. Each data point was determined from 15–30 ionic current time traces for individual captured complexes; each time trace had a duration of 5–10 s. (b–d) Plots of log(survival probability) vs dwell time for the lower amplitude level for complexes formed between wild-type Φ29 DNAP and DNA1-H_H, captured in the presence of (b) 0 μM dGTP or rGTP, (c) 5 μM dGTP, or (d) 1200 μM rGTP. In panels b–d, the dashed red line represents an exponential distribution with rate r2, which is the constant slope of log(survival probability) at 0 μM dGTP or rGTP and which is obtained by fitting to the data in Figure panel b (see text). The solid red fitting line in panel c shows the fit of the data to a model of two exponential modes. The dashed black fitting line in panel d shows the fit of the data to an exponential distribution. The dwell time samples were extracted from data files collected when complexes were captured at 180 mV; each file yields ∼8000–80000 dwell time samples for each amplitude level. In the plots, while 1 out of every 20 points is shown, the curves are fit to the full set of dwell time samples.
