Abstract
CTLA-4 is a costimulatory molecule that negatively regulates T cell activation. Originally identified in murine CD8+ T cells, it has been found to be rapidly induced on human T cells. Furthermore, CTLA-4 is expressed on regulatory T cells (Tregs). Clinically, targeting CTLA-4 has clinical utility in the treatment of melanoma. Whether the expression of CTLA-4 is differentially regulated in CD8+ vs. CD4+ human T cells is unclear. Here we analyzed CTLA-4 in normal human CD4+ and CD8+ T cell subsets and show for the first time that CTLA-4 is expressed significantly higher in the CD4+ T cells than in CD8+ T cells. CTLA-4 is higher at the protein and the transcriptional level in CD4+ T cells. This increase is due to activation of the CTLA-4 promoter, which undergoes acetylation at the proximal promoter. Furthermore, we show that blocking CTLA-4 on CD4+ T cells permits greater proliferation in CD4+ vs. CD8+ cells. These findings demonstrate a differential regulation of CTLA-4 on CD4+ and CD8+ T cell subsets, which is likely important to the clinical efficacy for anti-CTLA-4 therapies. The findings hint to strategies to modulate CTLA-4 expression by targeting epigenetic transcription to alter the immune response.
Keywords: CTLA-4, acetylation, CD4 T cells, NFAT
Introduction
Cytotoxic T lymphocyte associated protein 4 (CTLA-4/CD152) is a member of the membrane-bound single V domain subfamily within the immunoglobulin superfamily.1 CTLA-4 has significant homology to CD28, a co-stimulatory molecule that also binds the B7 ligands B7-1 (CD80) and B7-2 (CD86).2, 3 Although CD28 is detected in resting as well as activated T cells, CTLA-4 is primarily found on activated T cells and regulatory T cells (Tregs).4 Whereas engagement of B7 with CD28 typically provides the “second signal” required for T cell activation after a “first signal” is transduced via the T cell receptor (TCR), engagement of B7 with CTLA-4 results in inhibition of T cell activation and T cell proliferation.3 In Tregs, the expression is constitutive and is critical to their suppressive function.5–8 The precise mechanism by which CTLA-4 regulates the immune response is complex and not fully understood. However, it has been observed to transmit a negative signal through association with the protein phosphatase 2A, which inhibits the activation of the TCR and subsequently inhibits early cytokine gene expression.9 In addition, CTLA-4 binds B7 500 to 2500× more strongly than CD28.10, 11 An extensive lattice-like zipper structure of consisting of bivalent CTLA-4 homodimers and bivalent B7 homodimers effectively excludes the binding of CD28 to B7, suggesting that competitive inhibition is a key mechanism.12 In CTLA-4 deficient mice, profound lymphoid proliferation and death at 4 weeks are observed, suggesting the CTLA-4 plays an important role in downregulation of the immune system.13 The apparent rescue of these mice with CTLA-4-Ig further confirms the important role of CTLA-4 in immune regulation.14
The association of total serum IgE levels to chromosome 2q33, where there are CTLA-4 polymorphisms, as well as the identification of CTLA-4 as a susceptibility gene for thyroid autoantibody production, links CTLA-4 to specific human immune diseases.15, 16 CTLA-4 has also been linked to diabetes and atopic disease, in addition to hypothyroidism and Grave’s disease, as suggested by the aforementioned link between CTLA-4 and thyroid autoantibody production.17–21 Furthermore, we have found elevated CTLA-4 in patients with cutaneous T cell lymphoma (CTCL), a malignancy of memory T cells in which cytokine expression is abnormal.22, 23
Given the apparent association of CTLA-4 expression with numerous immune mediated diseases which furthermore is associated with specific T cell subsets, assessing the expression of CTLA-4 in different T cell subsets may provide insight into disease pathogenesis. Although CTLA-4 has been reported to be expressed at similar levels in CD4+ and CD8+ T cells in murine models, CTLA-4 expression in primary human peripheral T cell subsets may warrant further investigation.24 CTLA-4 is known to be differentially regulated in certain T cell subsets. In Tregs, CTLA-4 is constitutively expressed, due partially to FoxP3 control of CTLA-4 transcription in a NFAT-dependent manner, suggesting that there may be cell-type specific regulation of CTLA-4.25–28 CTLA-4 is also expressed at high levels on Th2 clones, both at the mRNA level and at the protein level.29 Furthermore, mutations in the promoter and untranslated regions of the CTLA-4 gene have resulted in decreased CTLA-4 expression in reporter gene assays, suggesting that transcriptional control of the CTLA-4 gene may also be essential to appropriate immune regulation.15 This suggests that agents that regulate gene expression via epigenetic mechanisms, such as histone deacetylase inhibitors, may be useful for modulating CTLA-4 expression in immunotherapy.
To better understand the regulation of CTLA-4, we studied its subset-specific expression in the context of CD4+ and CD8+ T cells. We show for the first time in human T cells that CTLA-4 is differentially expressed between CD4+ and CD8+ T cells. In T cells from normal individuals, there is preferential increase in CTLA-4 expression in CD4+ T cells, both at the cell surface and at the total protein level upon stimulation, but not in comparison to CD8+ T cells. Interferon, a cytokine important in cytotoxic T cells is higher in CD8+ than in CD4+ T cells. CTLA-4 is regulated at the level of transcription,28 and we observed that increased expression of CTLA-4 in CD4+ was associated with activation of the chromatin by the presence of acetylated histone H3 as well as NFAT1 binding to the CTLA-4 promoter. Finally, we demonstrate that the CD4+ bias in CTLA-4 expression affects CD4+ T cells by preferential suppression of CD4+ proliferation. Thus, in human T cells, there is increased expression of CTLA-4 in CD4+ T cells, which appears to be important in controlling their proliferation. This suggests that targeting CTLA-4 preferentially affects the function of the CD4+ T cell subset. These findings have implications in the clinical efficacy of anti-CTLA-4 therapies.
Results
Activated CD4+ T cells preferentially express CTLA-4
Although CTLA-4 was initially discovered in murine CD8+ T cells, whether there is a similar ability to express CTLA-4 among CD4+ and CD8+ T cells is unknown. The level of CTLA-4 induction is variable in PBMCs, and most human T cells do not express CTLA-4 in the resting state.4 To study whether differential control of inducible CTLA-4 expression could be observed in normal T cell subsets, we measured the level of CTLA-4 in human PBMCs after stimulation with PMA and A23187, strong activators of T cell gene expression.28 By flow cytometry analysis, we have previously shown that CTLA-4 was restricted to the CD3+ T cells in response to PMA/A23187.28 We then determined which subset of T cells was responsible for this expression. Because surface CD4 is down regulated upon stimulation with PMA in human T cells, we used CD8 as a marker to delineate CD8+ and CD8− subsets using 2-color flow cytometry.30 Surface CTLA-4 was detected in CD8− but not CD8+ T cell subsets after stimulation with PMA/A23187 (Figure 1a), suggesting that CD4+ T cells preferentially expressed CTLA-4 after activation.
Figure 1. CTLA-4 is preferentially induced in CD4 vs. CD8 T cells.
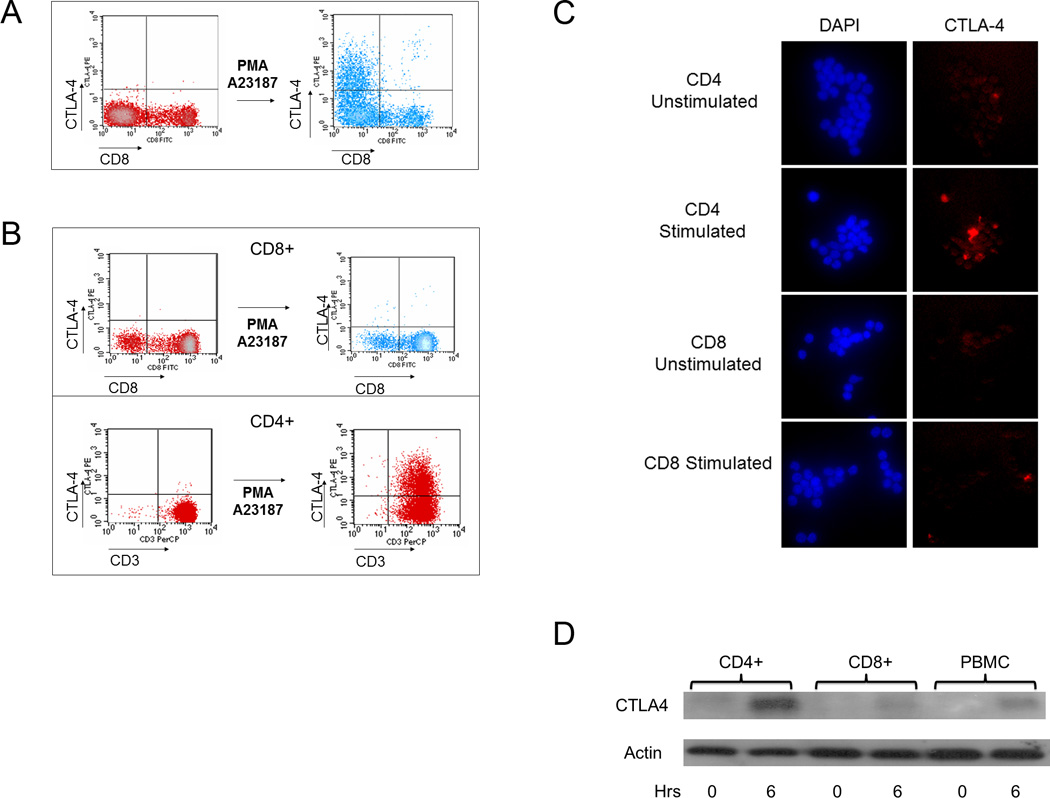
(a) The level of CTLA-4 expression was measured by flow cytometry before and after stimulation with PMA/A23187 as described in the Materials and Methods. Few CD8+ T cells express CTLA-4, suggesting that CTLA-4 is expressed mainly in non-CD8+ T cells. The results are representative of findings from three normal volunteers. (b) CD4 and CD8 T cells were purified using negative selection as described in the Materials and Methods. After stimulation with PMA/A23187, CTLA-4 expression was measured. CTLA-4 was minimal on the purified CD8+ subset (top panel) but was detected on the purified CD4+ subset (bottom panel). The results in each panel are representative of findings from three normal volunteers. (c) CTLA-4 is preferentially increased in stimulated CD4+ vs. CD8+ cells as assessed by immunofluorescence. Purified cells were stimulated with PMA/A23187 and fixed as described. Cells were stained with DAPI and anti-CTLA-4-PE as described in the Materials and Methods. Results are representative of three independent experiments. d) CD4+ and CD8+ T cells as well as bulk PBMCs were analyzed for total CTLA-4 expression. 10 million of each purified cell type was used in conditions following stimulation by PMA/A23187 as indicated. Following treatment with PMA/A23187 for 0 or 6 hours (horizontal axis), total protein was isolated as described in the Materials and Methods. 10 ug of total protein was separated by SDS-PAGE, transferred to PVDF membrane and immunoblotted for total CTLA-4 levels using anti-CTLA-4 followed by visualization by chemiluminescence.
To confirm that CTLA-4 is preferentially expressed on CD4+ T cells, we used negative selection to purify CD4+ and CD8+ populations from PBMCs, stimulated with PMA/A23187 and then analyzed them for CTLA-4 expression. Flow cytometry using CD3 as a marker showed that CTLA-4 was increased consistently higher in CD4+ purified T cells (Figure 1b, bottom panel) than in CD8+ purified T cells (Figure 1b, top panel) from the same individual. Consistent with results from the whole PMBC populations (Figure 1a), the CD8+ population did not express CTLA-4 efficiently upon stimulation (Figure 1b, top panel), whereas the CD4+ T cells rapidly expressed CTLA-4 upon stimulation (Figure 1b, bottom panel). Immunofluorescence of stimulated CD4 and CD8 T cells confirmed increased detection of CTLA-4 in stimulated CD4 T cells compared to CD8 T cells (Figure 1c).
To determine whether there is a difference in the total CTLA-4 protein between CD4+ and CD8+ T cells, total cell lysates were isolated from purified CD4+ and CD8+ T cells as well as bulk PBMCs, before and after stimulation, and immunoblots were performed to assess the total amount of CTLA-4 (Figure 1d). Minimal CTLA-4 was detectable in unstimulated cell populations (first, third, and fifth lanes from left). However, a high level of CTLA-4 protein was detected in stimulated CD4+ T cells (second lane from left), compared to stimulated CD8+ T cells (fourth lane from left) and bulk PBMCs (rightmost lane) after 6 hours of stimulation with PMA/A23187, indicating that total CTLA-4 expression is preferentially higher in CD4+ T cells after activation. These data suggest that upregulation in CTLA-4 expression is preferentially observed in CD4+ T cells is de novo and dependent upon stimulation with PMA/A23187.
CTLA-4 mRNA is differentially upregulated in CD4+ vs. CD8+ T cells
We surmised that increased CTLA-4 gene expression in CD4+ vs. CD8+ T cells could help explain the observed increase in CTLA-4 protein expression in CD4+ vs. CD8+ T cells. To test the hypothesis that increased CTLA-4 gene expression could be preferentially found in CD4+ vs. CD8+ T cells, we purified CD4+ and CD8+ T cell subsets via negative selection and then stimulated the purified cells to induce the expression of CTLA-4 protein. By quantitative RT-PCR, relative CTLA-4 expression was assessed (Figure 2a) and could be rapidly detected in CD4+ cells at one hour after stimulation, and persisted at 18 hours after stimulation (left panel, diamond hashed bars). In contrast, the relative CTLA-4 expression in CD8+ cells was minimally increased from 2 to 18 hours after stimulation (right panel, solid bars).
Figure 2. CTLA-4 gene expression is preferentially increased in CD4 T cells.
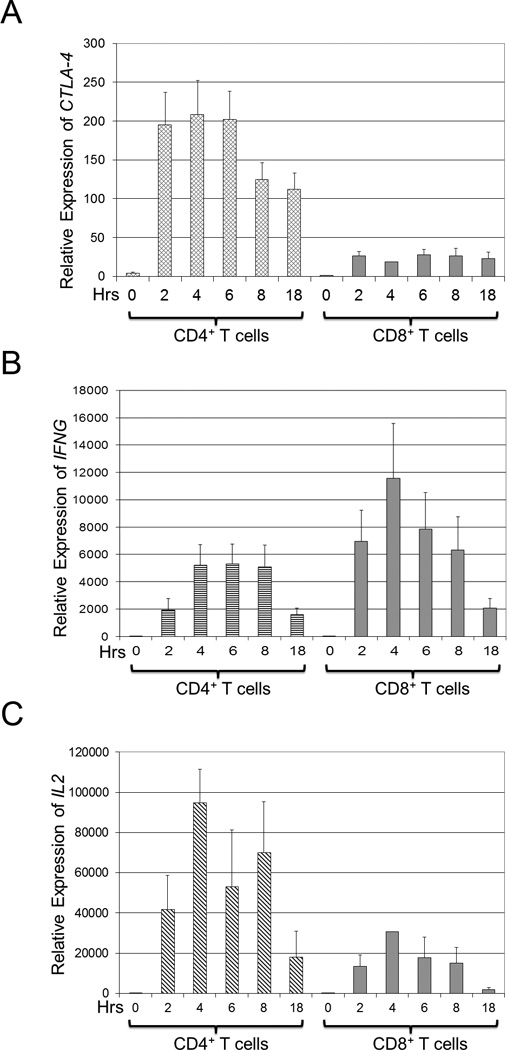
CD4+ and CD8+ T cells were isolated by negative selection from PBMCs as described in the Materials and Methods. Equal numbers of purified cells were stimulated with PMA/A23187 for 0, 2, 4, 6, 8, or 18 hours (hrs, horizontal axis), and total RNA was isolated and analyzed by qRT-PCR as described above. Results are depicted as average values ± s.d. of three normal volunteers. (a) The relative expression of CTLA-4 is higher in CD4+ compared to CD8+ T cells. CTLA-4 gene expression was normalized to CTLA-4 gene expression in unstimulated CD4+ T cells. (b) The relative expression of IFN-γ (IFNG) is higher in CD8+ compared to CD4+ T cells. IFNG gene expression was normalized to IFNG gene expression in unstimulated CD4+ T cells. (c) The relative expression of IL-2 (IL2) is higher in CD4+ compared to CD8+ T cells. IL2 gene expression was normalized to IL-2 gene expression in unstimulated CD4+ T cells.
To ensure that CD8+ T cells were activated, the expression of IFNG, the gene coding for interferon-γ, a cytokine preferentially expressed by activated CD8+ T cells,31 was analyzed (Figure 2b). By qRT-PCR, IFNG was higher in the CD8+ T cells (right panel, solid bars) than in CD4+ T cells (left panel, horizontal hash bars), indicating that the CD8+ T cells were functional and adequately stimulated. To confirm that CD4+ T cells were activated, the expression of IL2, a cytokine preferentially expressed by activated CD4+ T cells,31 was analyzed (Figure 2c) and found by qRT-PCR to be higher in CD4+ T cells (left panel, slanted hash bars) than in CD8+ T cells (right panel, solid bars). These findings show that the CTLA-4 gene is differentially expressed CD4 vs. CD8 in activated T cells.
The CTLA-4 promoter is preferentially associated with increased NFAT1 and acetylated histones in CD4+ compared to CD8+ T cells
We have previously shown via EMSA and ChIP assays from human T cells that CTLA-4 transcription is dependent upon NFAT1 binding to the proximal promoter of the CTLA-4 gene.28 This is consistent with a prior report that NFAT1 levels are increased in primary human CD4 T cells primed in vitro.32 We hypothesized that higher binding of NFAT1 to the CTLA-4 promoter in CD4+ cells vs. CD8+ cells could be a mechanism to explain the higher transcriptional expression of CTLA-4 in CD4+ vs. CD8+ T cells. Therefore, to determine whether a greater level of NFAT1 activity could be found at the CTLA-4 promoter in CD4+ vs. CD8+ T cells, EMSA assays were performed as previously described (Figure 3a).28 Using the NFAT1 binding site from the CTLA-4 promoter (*C(−280)NFAT) as a probe, we found that both unstimulated and stimulated CD4 cells had higher levels of NFAT activity in CD4+ vs. CD8+ cells as evidenced by upward shifts/lower motility of the NFAT-(*C(−280)NFAT)) complexes in lysates derived from CD4+ cells (first and fifth lanes from the left) vs. CD8+ cells (third and seventh lanes from the left). However, lysates from stimulated CD4+ cells expressed the highest level of DNA binding (fifth lane from the left), consistent with increased NFAT transcription in CD4 cells after stimulation. After incubation of the cell lysates with unlabeled probe which competed with the radiolabelled probe (second, fourth, sixth, and eight lanes from the left), minimal radiolabelled probe was detected, confirming specificity of the radiolabelled probe to NFAT.
Figure 3. Increased NFAT activity is found in CD4+ vs. CD8+ T cells at the CTLA-4 promoter.
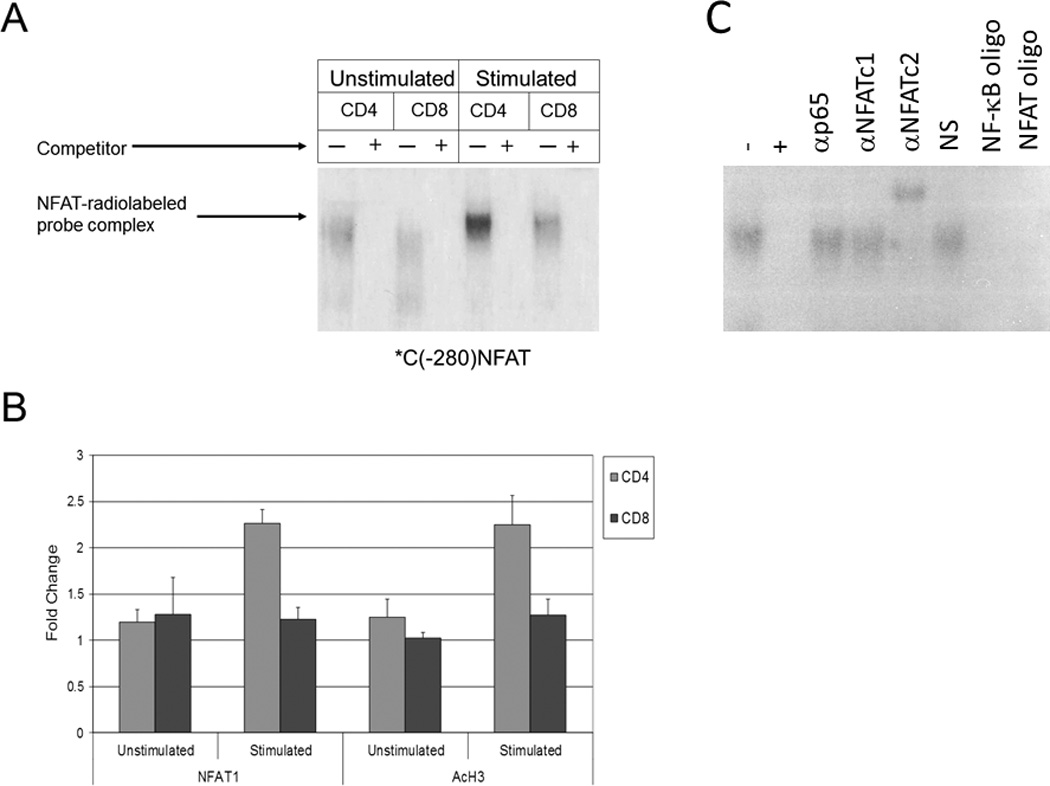
a) Protein extracts from unstimulated and stimulated CD4+ and CD8+ T cells were incubated with a radiolabelled probe representing an NFAT binding site in the CTLA-4 promoter (*C(−280)NFAT). Subsequently, mixtures were then incubated with or without excess competitor unlabeled probe (+ or −) and NFAT1 binding was assayed via 4% polyacrylamide gel electrophoresis. b) CHIP assays were performed on the CTLA-4 promoter region using anti-NFAT1 (left panel) and anti-acetylated histone H3 (AcH3, right panel) antibodies in unstimulated and stimulated CD4+ and CD8+ T cells. Normalization for equal cell numbers was 10% of the input DNA. Results in each panel are averages from two independent experiments. c) Gel shift mobility assay (EMSA) was performed as described above with our without excess competitor unlabeled probe (+ or −) and gel shift was assessed using antibodies to the p65 subunit of NF-κB, NFAT1, and NFAT2. Normal serum (NS), NF-κb oligonucleotides (NF-κb oligo) and NFAT oligonucleotides (NFAT oligo) were used as additional controls.
To study the activity of the CTLA-4 promoter in CD4+ and CD8+ T cells, purified CD4+ and CD8+ T cells were stimulated, DNA was extracted for use in ChIP assays. To confirm that more NFAT is present at the proximal CTLA-4 promoter in CD4+ vs. CD8+ T cells, ChIP studies were performed with anti-NFAT1 and anti-acetylated histone H3 (AcH3) antibodies (Figure 3b) using purified T cells were stimulated with PMA/A23187 for 6 hours prior to fixation and sonication. Our studies showed a corresponding higher level of NFAT1 at the CTLA-4 promoter in CD4+ vs. CD8+ T cells (left panel). Furthermore, the use of an anti-acetylated histone H3 antibody to measure transcriptionally active chromatin revealed a higher level of acetylated histone H3 at the CTLA-4 promoter in CD4+ T cells compared to CD8+ T cells, further supporting our finding of higher transcription of the CTLA-4 gene in CD4+ vs. CD8+ T cells (right panel).
It was noted that the observed difference in NFAT1 binding to the CTLA-4 promoter in vivo (ChIP) following stimulation (Figure 3a), while appreciable, did appear somewhat modest. Prior studies regarding NFAT2 have demonstrated that it is upregulated upon CD4 T cell stimulation and that NFAT1 expression contributes to the subsequent induction of NFAT2 via the binding of NFAT1 to the NFAT2 promoter during T cell activation.32–34 We have previously demonstrated that NFAT2 does not bind the NFAT consensus region in the CTLA-4 promoter using an NFAT probe centered around −280 bp in an EMSA assay.28 These findings were reconfirmed in an EMSA assay which also demonstrated that the p65 subunit of NF-kB does not bind *C(−280)NFAT (Figure 3c).
NFAT1 is differentially expressed in stimulated CD4+ vs. CD8+ cells
To determine whether the difference in the expression of CTLA-4 between CD4+ and CD8+ T cell subsets was due to differential expression of NFAT1 protein, the level of NFAT1 was measured in CD4+ and CD8+ T cells. Total protein was isolated from purified and stimulated CD4+ and CD8+ T cells and immunoblotted using an anti-NFAT1 antibody (Figure 4a). We found a low level of NFAT1 in unstimulated CD4+ and CD8+ cells (first and third lanes from the left). However in stimulated CD4+ T cells, there is a significantly greater level of NFAT1 protein (fourth lane from the left) in CD4+ compared to stimulated CD8+ T cells (second lane from the left). These results were confirmed by qRT-PCR (Figure 4b). Stimulated CD4+ cells (second column from left) expressed higher levels of normalized NFAT1 than stimulated CD8+ cells (fourth column from left), and both of these populations expressed higher levels of normalized NFAT1 compared to their respective unstimulated populations (first and third columns from left). These results confirm that NFAT1 expression is higher at both the protein and transcriptional level in stimulated CD4+ vs. CD8+ T cells and may contribute to the increased levels of CTLA-4 expression seen in CD4+ vs. CD8+ T cells.
Figure 4. NFAT1 is increased in CD4+ T cells compared to CD8+ T cells.
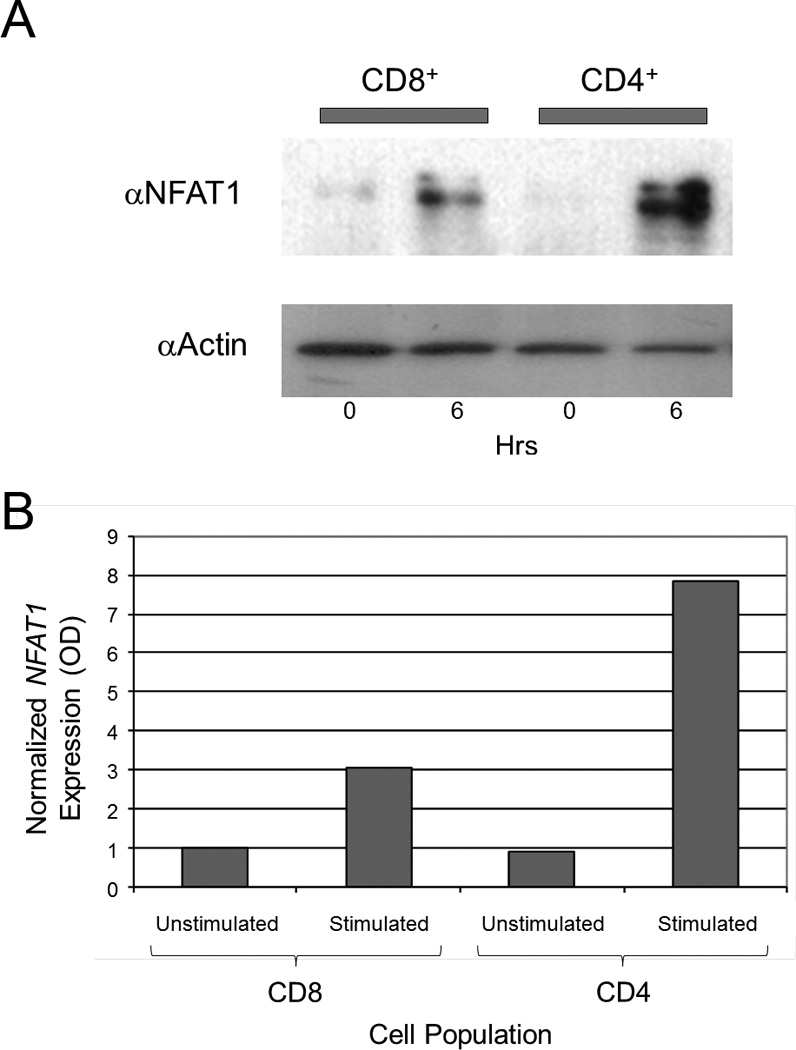
(a) NFAT1 is increased in stimulated CD4+ cells vs. stimulated CD8+ cells by immunoblot. Total protein was isolated from purified unstimulated (0 hours, horizontal axis) and stimulated (6 hours, horizontal axis) CD4+ and CD8+ T cells. Equal amount amounts of protein (10 µg) was analyzed by SDS-PAGE and immunoblotted against NFAT1 with an anti-NFAT1 antibody (αNFATc2) as shown. The level of actin was determined by immunoblotting the same membrane with an anti-actin antibody (αActin) after stripping the anti-NFAT1 to control for equal protein loading. Results are representative of three independent experiments. (b) NFAT1 expression was measured by qRT-PCR and normalized to β2-microglobulin. Stimulated CD4+ cells show higher relative expression of NFAT1 compared to stimulated CD8+ cells.
Increased CTLA-4 on CD4+ T cells restricts CD4+ T cell proliferation
Anti-CTLA-4 has been shown to block interaction between B7 and CTLA-4, thereby removing inhibitory regulation of signaling.35, 36 To determine the functional significance of the differential expression of CTLA-4 on CD4+ vs. CD8+ T cells, we performed a proliferation assay using CD4+ and CD8+ T cells in the presence or absence of anti-CTLA-4 blocking antibodies (Figure 5). When anti-CTLA-4 mAb was added to a mixed-lymphocyte reaction (MLR) with CD4+ T cells, there was a measurable enhancement in the proliferation of CD4+ T cells. In contrast, the addition of anti-CTLA-4 antibodies to CD8+ T cells in an MLR did not significantly affect their proliferation.
Figure 5. CTLA-4 expression on CD4+ T cells reduces the proliferative potential of CD4+ T cells.
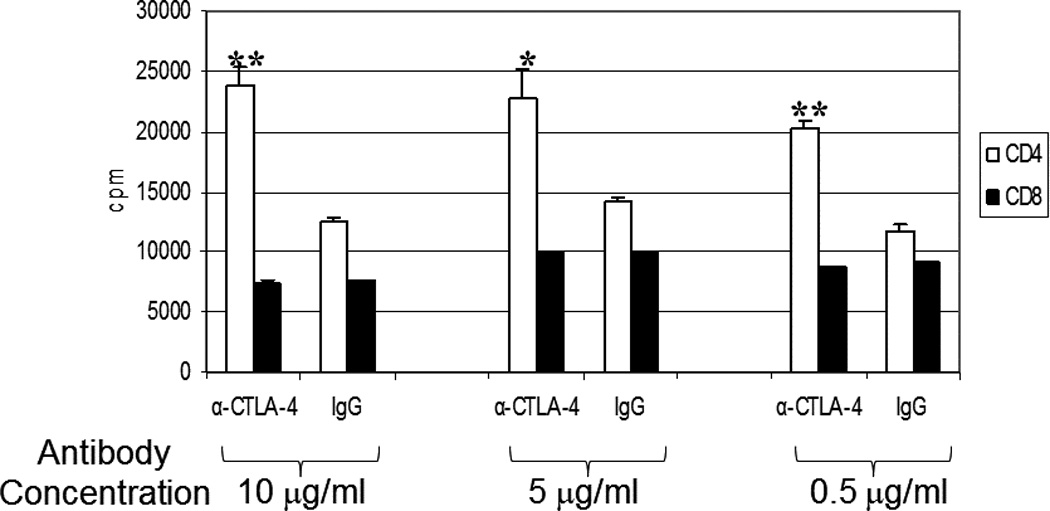
Purified CD4+ or CD8+ T cells were cultured as described in the Materials and Methods. Mixed lymphocyte reaction was performed in the presence of anti-CTLA-4 or IgG isotype control antibody was titrated at 10 µg/ml, 5 µg/ml, or 0.5 µg/ml with CD4+ or CD8+ T cells as indicated over 7 days followed by additional of H3-thymidine. Average cpm were calculated ± s.d. p values were calculated comparing the difference in cpm proliferation in CD4+ vs. CD8+ T cells and were < 0.01 (*) or < 0.001 (**) as indicated. Results are representative of 2 independent experiments.
Discussion
Although the functional role of CTLA-4 on T cell inhibition in the context of CD4 cells and regulatory T cells (Tregs) are well described, the differential expression of CTLA-4 in human T cell subsets in non-CD4 T cells has not been fully described.5, 7, 37–41 Moreover the potency of antibodies that target CTLA-4 in treating cancer suggest that thorough characterization the expression of CTLA-4 in different T cells is essential to understanding the target population and the mechanism of response. In mice, CD8+ T cells express CTLA-4 when activated.35, 36, 42, 43 However, it is unclear what significance the absence of CTLA-4 on these cells might portend, as is the case in our study with activated human CD8+ T cells. One study in mice has shown that autoimmune CD8+ T cell activity in CTLA-4 knockout mice also containing transgenic Vβ13 TCR chains is only manifested when CD4+ T cells lacking CTLA-4 are also present, suggesting that the absence of CTLA-4 on CD8+ cells is insufficient for an autoimmune process to develop in this system and that it may be the absence of CTLA-4 on CD4+ T cells that enables autoimmunity to develop.44 Furthermore, CTLA-4-blocking antibodies have been shown in mice to increase the number of antigen-specific CD8+ memory T cells capable of producing IFN-γ and TNF-α45. It has been shown that anti-CTLA-4 increases IL-2 production by CD4+ helper T cells and that IL-2 signaling can increase terminal differentiation of CD8+ T cells, thereby providing further evidence of the importance of CTLA-4 on CD4+ T cells in terms of influencing changes in CD8+ T cell activity.45–47 From these murine studies, it appears that the critical functions of CTLA-4 are primarily mediated by its presence on CD4+ T cells rather than via its apparent presence on CD8+ T cells.
Based upon these murine studies, we hypothesized that a similar bias in CTLA-4 function toward CD4+ vs. CD8+ T cells might also be apparent in humans. Limited studies have assessed the intracellular expression of CTLA-4 on CD8+ human T cells, showing that in normal unstimulated cells, there is no appreciable intracellular CTLA-4 in CD4+ or CD8+ cells.48 Another study has looked at differential expression of CTLA-4 T cell subsets, but this study only examined purified CD4+ human T cells, confirming low intracellular expression of CTLA-4 in naïve CD4+ T cells and higher expression in CD4+CD25+ Tregs.49 Therefore, the characterization of CTLA-4 in unstimulated and stimulated CD8+ T cells relative to CD4+ T cells has not been yet reported.
While studying the regulation of CTLA-4 induction by PMA/A23187, we observed that only a fraction of T cells in the normal peripheral blood have the ability to express CTLA-4 and thus reasoned that the low level of CTLA-4 expression might reflect a heterogeneity of T cell subsets. Our results showing that the expression of CTLA-4 is preferentially expressed in the CD4+ T cell subset and is absent on CD8+ T cells appears to correlate with the important role that CTLA-4 apparently plays in CD4+ T cell-based regulation of CD8+ T cells in the mouse even though CTLA-4 was initially cloned from the CD8+ T cells in mice.45–47, 50
Our results show that, in humans, CTLA-4 is expressed and regulated differentially between CD4+ and CD8+ T cells. This difference in CTLA-4 expression can be detected at the total protein level and via differences in the RNA expression of the CTLA-4 gene. Our demonstration of found higher levels of NFAT1 in CD4+ vs. CD8+ T cells correlating with respectively increased CTLA-4 expression from the same individual suggests a mechanism to at least partially account for the increased levels of CTLA-4 in CD4+ vs. CD8+ T cells.
Our analysis of NFAT1 at the CTLA-4 promoter using ChIP analysis builds upon our previously published work by showing that NFAT1 activity is preferentially increased at the CTLA-4 promoter in CD4+ vs. CD8+ cells.28 Defective phosphorylation of the NFAT transactivational domain can be observed in antigen-stimulated CD8+ T cells compared to CD4+ T cells, suggesting a mechanism for at least partially explaining the lower NFAT activity in CD8+ T cells vs. CD4+ T cells.51 Furthermore, our work demonstrating that the acetylation status of the chromatin of the CTLA-4 gene of CD4+ T cells differs fundamentally from that in CD8+ T cells sheds further light upon a mechanism for why there is differential expression of CTLA-4 in human CD4+ vs. CD8+ T cells.
The increase in CTLA-4 on CD4+ T cells is functionally significant, and blockade of CTLA-4 enhances proliferation of the CD4+ T cell subset. This corresponding finding is not seen in the CD8+ T cell subset and is likely because the latter cells did not express detectable levels of CTLA-4 upon stimulation in the first place. This highlights the importance of CD4+ T cells as the principle mediators of CTLA-4 function and may give us further insight into how anti-CTLA-4 antibodies such as ipilimumab can boost the immune system response to cancer in humans.52–54
Currently, anti-CTLA-4 antibodies such as ipilimumab have been shown to offer survival benefit in patients with melanoma.52–55 It is now apparent that treatment of patients with ipilimumab can increase activated and antigen-specific CD4+ and CD8+ T cells with a decrease in naïve CD4+ and CD8+ T cells.56, 57 Our studies now provide further insight into why CD4+ T cells, owing to their high expression of CTLA-4, relative to CD8+ T cells, may be the principle mediators of CTLA-4 function in humans, mirroring the results seen in mice.45–47, 50 Furthermore, the increased expression of CTLA-4 on CD4+CD25hi Tregs, suggests that mechanisms that we have described above may be also more apparent in CD4+CD25hi Tregs vs. bulk CD4+ T cell populations.5, 7
Our proliferation studies also demonstrate that increased expression of CTLA-4 in the human CD4+ T cell population may be required to control proliferation. As our studies have shown that human CD8+ T cells do not express CTLA-4 upon stimulation but proliferate, albeit not as well as human CD4+ T cells treated with anti-CTLA-4, the presence of CTLA-4 on CD4+ T cells may in vivo be necessary for preventing uncontrolled CD8+ and CD4+ effector function and the prevention of autoimmunity. These findings are consistent with the phenotype seen in murine CTLA-4−/− knock out animals in which there is profound lymphoproliferation, predominantly of CD4+ T cells58. Loss of CTLA-4 leads to non-malignant expansion of the CD4 T cell subset.58 These observations indicate that CD4+ T cells have an inherent difference in the potential to proliferate, relative to CD8+ T cells and the increased expression of CTLA-4 may be a mechanism for controlling cell division and perhaps, evolutionally, why CTLA-4 expression is biased towards CD4+ T cells.
In summary, the preferential expression of CTLA-4 on CD4 T cells, mediated by increased NFAT1, may have implications in targeting CTLA-4 for therapy. The preferential regulation of the CD4+ T cells potentially leaves CD8+ T cells free to remain unmanipulated and unopposed.
Materials and Methods
Patients & cell preparation
A protocol for the acquisition of human blood was approved by the Henry Ford Hospital Institutional Review Board (HFHIRB). Human blood buffy coat specimens were either obtained from residual pheresis purification units (gifts from the American Red Cross of the Detroit) or from the blood of normal healthy volunteers who were provided written informed consent for this research study under HFHIRB-approved protocols abiding by the guidelines set forth by the Declaration of Helsinki. Peripheral blood mononuclear cells (PBMCs) were subsequently purified using a ficoll gradient as previously described.28 Prior studies regarding the induction of CTLA-4 showed that both CD4 and CD8 cell populations could express CTLA-4 and that it could be expressed via either CD3/CD28 activation or via phorbol myristate acetate (PMA) plus calcium ionophore.59, 60 We have previously demonstrated CTLA-4 induction in T cells after stimulation with PMA at 25 ng/ml and A23187 calcium ionophore at 0.1 µg/ml for 18 hours.28 Therefore, stimulation of these cells was performed as previously described by our group.28 For the purification of CD4+ or CD8+ T cells, a magnetic bead/column-based purification kit from Stemcell Technologies (Vancouver, BC, Canada) was utilized, and the manufacturer’s instructions were followed. The enrichment of CD4+ cells and CD8+ cells were done by negative selection to prevent premature activation of the cells of interests. The purity of CD4+ cells and CD8+ cells was tested by staining purified cells with the appropriate fluorescent antibodies for analysis. Purity of the CD4+ and CD8+ cells were found to be greater than 95% and 85% respectively.
Antibodies
Anti-actin antibody, anti- anti-NFATc2 (NFAT1) were purchased from Santa Cruz Biotechnologies (Santa Cruz, CA, USA). Anti-acetylated histone H3 was purchased from Upstate Biotechnology (Billerica, MA, USA).
Biochemicals
Lymphoprep (Nycomed: Roskilde, Denmark); CsA, PMA, poly (dIdC) (Sigma-Aldrich: St. Louis, MO, USA); A23187 calcium ionophore, 11R-VIVIT (EMD Biosciences: Darmstadt, Germany); TRIzol, oligonucleotides, RPMI (Invitrogen: Carlsbad, CA, USA); Supersignal (Pearce Biotechnology, Inc.: Rockford, IL, USA)
Quantitative PCR analysis
Total RNA was isolated from PBMCs using TRIzol as recommended by the manufacturer. As previously described, reverse transcription was performed with total RNA to generate cDNA, and quantitative PCR was performed.28 Primers for analysis of CTLA-4 (−380 5’-ATTGGGATTTAGGAGGACCC-3’; −330 5’-CCACTTAGTTATCCAGATC CTC-3’), β2-microglobulin (5’-TCTACTTTGAGTGCTGTCTCCATGT-3’ and 5’-AAGTTGCCAGCCCTCCTAGAG-3’), and interferon (5’-tcctgtcactgtctcacttaatcctt-3’ and 5’-ttaggttggctgcctagttgg-3’) have similar amplification efficiencies. Analysis of relative gene expressions was performed using the 2−ΔΔCT method.61 Assessment of CTLA-4 expression in each clinical sample was performed in duplicate, and results were normalized in each sample relative to the level of β2-microglobulin.
Immunoblot Analysis
Protein samples were prepared from whole cell extracts as previously described.28 Immunoblotting was also performed as previously described.28
Chromatin immunoprecipitation assay (ChIP)
ChIP was performed as previously described and adapted from a method described by the Farnham group.28, 62 PCR was performed using primers (−380 5’-ATTGGGATTTAGGAGGACCC-3’; −330 5’-CCACTTAGTTATCCAGATC CTC-3’) for the proximal CTLA-4 promoter to determine the level of DNA precipitated using antibodies.
Electrophoretic mobility shift assay (EMSA)
EMSA was performed as previously described28. In brief, a γ-32P-labeled dsC-(−280)NFAT probe representing the NFAT binding site −280 bp from the CTLA-4 transcription start site (*C(−280)NFAT) was used for the assessment of gel shifts. Protein extracts from unstimulated and stimulated lymphocyte were incubated with radiolabelled probe, and the mobility of the probes was assessed on a 4% nondenaturing polyacrylamide gels as previously described.28 As a control, excess unlabelled probe was also used to compete off the labeled probe, as previously described.28
Lymphocyte proliferation assay
PBMCs were prepared from two normal individuals for each proliferation study. Stimulator PBMCs were irradiated with 2000 Rads and cultured with responders consisting of either CD4+ or CD8+ cells in a culture medium containing RPMI-1640 with 10% heat inactivated bovine serum (FBS), 2 mM glutamine, 1% Penicillin/Streptomycin (100U/ml) for 6 days. Irradiated stimulator cells were cultured with 105 responder cells at a ratio of 1:1 and 10:1 individual wells of a 96 well round bottom plate and a total volume of 200 ul per well. Subsequently, 3H incorporation was assessed at the last 16 hours of culture. To study the effect of CTLA-4 inhibition, human anti-CTLA-4 or anti-IgG was added to cultures at a final concentration of 10 ug/ml, 5 ug/ml, and 0.5ug/ml. Irradiated stimulator PBMCs and responder cells plated alone did not show any proliferative activity.
Statistical analysis
Statistical analysis was performed using Microsoft Excel software.
Acknowledgements
This work was supported in part by funds from the National Cancer Institute P30 supplement to the OSU Comprehensive Cancer Center (PI M. Caligiuri), funds to Henry Ford Hospital, and OSU Dermatology Research funds.
Footnotes
Conflict of Interest Disclosure
None of the authors have any competing financial interests to declare in relation to the work described in this manuscript.
References
- 1.Dariavach P, Mattéi M-G, Golstein P, Lefranc M-P. Human Ig superfamily CTLA-4 gene: chromosomal localization and identity of protein sequence between murine and human CTLA-4 cytoplasmic domains. European Journal of Immunology. 1988;18(12):1901–1905. doi: 10.1002/eji.1830181206. [DOI] [PubMed] [Google Scholar]
- 2.Harper K, Balzano C, Rouvier E, Mattei M, Luciani M, Golstein P. CTLA-4 and CD28 activated lymphocyte molecules are closely related in both mouse and human as to sequence, message expression, gene structure, and chromosomal location. The Journal of Immunology. 1991;147(3):1037–1044. [PubMed] [Google Scholar]
- 3.Auchincloss H, Turka LA. CTLA-4: not all costimulation is stimulatory. J Immunol. 2011;187(7):3457–3458. doi: 10.4049/jimmunol.1102316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lindsten T, Lee K, Harris E, Petryniak B, Craighead N, Reynolds P, et al. Characterization of CTLA-4 structure and expression on human T cells. The Journal of Immunology. 1993;151(7):3489–3499. [PubMed] [Google Scholar]
- 5.Friedline RH, Brown DS, Nguyen H, Kornfeld H, Lee J, Zhang Y, et al. CD4+ regulatory T cells require CTLA-4 for the maintenance of systemic tolerance. J Exp Med. 2009;206(2):421–434. doi: 10.1084/jem.20081811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schmidt EM, Wang CJ, Ryan GA, Clough LE, Qureshi OS, Goodall M, et al. Ctla-4 controls regulatory T cell peripheral homeostasis and is required for suppression of pancreatic islet autoimmunity. J Immunol. 2009;182(1):274–282. doi: 10.4049/jimmunol.182.1.274. [DOI] [PubMed] [Google Scholar]
- 7.Wing K, Onishi Y, Prieto-Martin P, Yamaguchi T, Miyara M, Fehervari Z, et al. CTLA-4 control over Foxp3+ regulatory T cell function. Science. 2008;322(5899):271–275. doi: 10.1126/science.1160062. [DOI] [PubMed] [Google Scholar]
- 8.Zheng SG, Wang JH, Stohl W, Kim KS, Gray JD, Horwitz DA. TGF-beta requires CTLA-4 early after T cell activation to induce FoxP3 and generate adaptive CD4+CD25+ regulatory cells. J Immunol. 2006;176(6):3321–3329. doi: 10.4049/jimmunol.176.6.3321. [DOI] [PubMed] [Google Scholar]
- 9.Chuang E, Fisher TS, Morgan RW, Robbins MD, Duerr JM, Vander Heiden MG, et al. The CD28 and CTLA-4 receptors associate with the serine/threonine phosphatase PP2A. Immunity. 2000;13(3):313–322. doi: 10.1016/s1074-7613(00)00031-5. [DOI] [PubMed] [Google Scholar]
- 10.Peggs KS, Quezada SA, Korman AJ, Allison JP. Principles and use of anti-CTLA4 antibody in human cancer immunotherapy. Curr Opin Immunol. 2006;18(2):206–213. doi: 10.1016/j.coi.2006.01.011. [DOI] [PubMed] [Google Scholar]
- 11.Wolchok JD, Saenger Y. The Mechanism of Anti-CTLA-4 Activity and the Negative Regulation of T-Cell Activation. The Oncologist. 2008;13(suppl 4):2–9. doi: 10.1634/theoncologist.13-S4-2. [DOI] [PubMed] [Google Scholar]
- 12.Stamper CC, Zhang Y, Tobin JF, Erbe DV, Ikemizu S, Davis SJ, et al. Crystal structure of the B7-1/CTLA-4 complex that inhibits human immune responses. Nature. 2001;410(6828):608–611. doi: 10.1038/35069118. [DOI] [PubMed] [Google Scholar]
- 13.Tivol EA, Borriello F, Schweitzer AN, Lynch WP, Bluestone JA, Sharpe AH. Loss of CTLA-4 leads to massive lymphoproliferation and fatal multiorgan tissue destruction, revealing a critical negative regulatory role of CTLA-4. Immunity. 1995;3(5):541–547. doi: 10.1016/1074-7613(95)90125-6. [DOI] [PubMed] [Google Scholar]
- 14.Tivol E, Boyd S, McKeon S, Borriello F, Nickerson P, Strom T, et al. CTLA4Ig prevents lymphoproliferation and fatal multiorgan tissue destruction in CTLA-4-deficient mice. The Journal of Immunology. 1997;158(11):5091–5094. [PubMed] [Google Scholar]
- 15.Howard TD, Postma DS, Hawkins GA, Koppelman GH, Zheng SL, Wysong AK, et al. Fine mapping of an IgE-controlling gene on chromosome 2q: Analysis of CTLA4 and CD28. J Allergy Clin Immunol. 2002;110(5):743–751. doi: 10.1067/mai.2002.128723. [DOI] [PubMed] [Google Scholar]
- 16.Tomer Y, Greenberg DA, Barbesino G, Concepcion E, Davies TF. CTLA-4 and Not CD28 Is a Susceptibility Gene for Thyroid Autoantibody Production. Journal of Clinical Endocrinology & Metabolism. 2001;86(4):1687–1693. doi: 10.1210/jcem.86.4.7372. [DOI] [PubMed] [Google Scholar]
- 17.Hizawa N, Yamaguchi E, Jinushi E, Konno S, Kawakami Y, Nishimura M. Increased total serum IgE levels in patients with asthma and promoter polymorphisms at CTLA4 and FCER1B. J Allergy Clin Immunol. 2001;108(1):74–79. doi: 10.1067/mai.2001.116119. [DOI] [PubMed] [Google Scholar]
- 18.Marron MP, Raffel LJ, Garchon HJ, Jacob CO, Serrano-Rios M, Martinez Larrad MT, et al. Insulin-dependent diabetes mellitus (IDDM) is associated with CTLA4 polymorphisms in multiple ethnic groups. Hum Mol Genet. 1997;6(8):1275–1282. doi: 10.1093/hmg/6.8.1275. [DOI] [PubMed] [Google Scholar]
- 19.Ueda H, Howson JM, Esposito L, Heward J, Snook H, Chamberlain G, et al. Association of the T-cell regulatory gene CTLA4 with susceptibility to autoimmune disease. Nature. 2003;423(6939):506–511. doi: 10.1038/nature01621. [DOI] [PubMed] [Google Scholar]
- 20.Nithiyananthan R, Heward JM, Allahabadia A, Franklyn JA, Gough SC. Polymorphism of the CTLA-4 gene is associated with autoimmune hypothyroidism in the United Kingdom. Thyroid. 2002;12(1):3–6. doi: 10.1089/105072502753451896. [DOI] [PubMed] [Google Scholar]
- 21.Kotsa K, Watson PF, Weetman AP. A CTLA-4 gene polymorphism is associated with both Graves disease and autoimmune hypothyroidism. Clin Endocrinol (Oxf) 1997;46(5):551–554. doi: 10.1046/j.1365-2265.1997.1710996.x. [DOI] [PubMed] [Google Scholar]
- 22.Wong HK, Wilson AJ, Gibson HM, Hafner MS, Hedgcock CJ, Berger CL, et al. Increased expression of ctla-4 in malignant T-cells from patients with mycosis fungoides -- cutaneous T cell lymphoma. J Invest Dermatol. 2006;126(1):212–219. doi: 10.1038/sj.jid.5700029. [DOI] [PubMed] [Google Scholar]
- 23.Chong BF, Wilson AJ, Gibson HM, Hafner MS, Luo Y, Hedgcock CJ, et al. Immune function abnormalities in peripheral blood mononuclear cell cytokine expression differentiates stages of cutaneous T-cell lymphoma/mycosis fungoides. Clin Cancer Res. 2008;14(3):646–653. doi: 10.1158/1078-0432.CCR-07-0610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Freeman GJ, Lombard DB, Gimmi CD, Brod SA, Lee K, Laning JC, et al. CTLA-4 and CD28 mRNA are coexpressed in most T cells after activation. Expression of CTLA-4 and CD28 mRNA does not correlate with the pattern of lymphokine production. J Immunol. 1992;149(12):3795–3801. [PubMed] [Google Scholar]
- 25.Takahashi T, Tagami T, Yamazaki S, Uede T, Shimizu J, Sakaguchi N, et al. Immunologic Self-Tolerance Maintained by Cd25+Cd4+Regulatory T Cells Constitutively Expressing Cytotoxic T Lymphocyte–Associated Antigen 4. The Journal of Experimental Medicine. 2000;192(2):303–310. doi: 10.1084/jem.192.2.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wu Y, Borde M, Heissmeyer V, Feuerer M, Lapan AD, Stroud JC, et al. FOXP3 Controls Regulatory T Cell Function through Cooperation with NFAT. Cell. 2006;126(2):375–387. doi: 10.1016/j.cell.2006.05.042. [DOI] [PubMed] [Google Scholar]
- 27.Jain N, Nguyen H, Chambers C, Kang J. Dual function of CTLA-4 in regulatory T cells and conventional T cells to prevent multiorgan autoimmunity. Proceedings of the National Academy of Sciences. 2010;107(4):1524–1528. doi: 10.1073/pnas.0910341107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gibson HM, Hedgcock CJ, Aufiero BM, Wilson AJ, Hafner MS, Tsokos GC, et al. Induction of the ctla-4 gene in human lymphocytes is dependent on NFAT binding the proximal promoter. J Immunol. 2007;179(6):3831–3840. doi: 10.4049/jimmunol.179.6.3831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Alegre ML, Shiels H, Thompson CB, Gajewski TF. Expression and function of CTLA-4 in Th1 and Th2 cells. J Immunol. 1998;161(7):3347–3356. [PubMed] [Google Scholar]
- 30.Weyand CM, Goronzy J, Fathman CG. Modulation of CD4 by antigenic activation. J Immunol. 1987;138(5):1351–1354. [PubMed] [Google Scholar]
- 31.Morvan PY, Picot C, Dejour R, Genetet B, Genetet N. Distinct pattern of IL-2 and IFN-gamma gene expression in CD4 and CD8 T cells: cytofluorometric analysis at a single cell level using non-radioactive probes. Cell Mol Biol (Noisy-le-grand) 1995;41(7):945–957. [PubMed] [Google Scholar]
- 32.Cron RQ, Bort SJ, Wang Y, Brunvand MW, Lewis DB. T Cell Priming Enhances IL-4 Gene Expression by Increasing Nuclear Factor of Activated T Cells. The Journal of Immunology. 1999;162(2):860–870. [PubMed] [Google Scholar]
- 33.Zhou B, Cron RQ, Wu B, Genin A, Wang Z, Liu S, et al. Regulation of the Murine Nfatc1 Gene by NFATc2. Journal of Biological Chemistry. 2002;277(12):10704–10711. doi: 10.1074/jbc.M107068200. [DOI] [PubMed] [Google Scholar]
- 34.Torgerson TR, Genin A, Chen C, Zhang M, Zhou B, Añover-Sombke S, et al. FOXP3 Inhibits Activation-Induced NFAT2 Expression in T Cells Thereby Limiting Effector Cytokine Expression. The Journal of Immunology. 2009;183(2):907–915. doi: 10.4049/jimmunol.0800216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Krummel MF, Allison JP. CD28 and CTLA-4 have opposing effects on the response of T cells to stimulation. J Exp Med. 1995;182(2):459–465. doi: 10.1084/jem.182.2.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Krummel MF, Allison JP. Pillars article: CD28 and CTLA-4 have opposing effects on the response of T cells to stimulation. J Immunol. 2011;187(7):3459–3465. The journal of experimental medicine. 1995. 182: 459–465. [PubMed] [Google Scholar]
- 37.Perkins D, Wang Z, Donovan C, He H, Mark D, Guan G, et al. Regulation of CTLA-4 expression during T cell activation. J Immunol. 1996;156(11):4154–4159. [PubMed] [Google Scholar]
- 38.Egen JG, Kuhns MS, Allison JP. CTLA-4: new insights into its biological function and use in tumor immunotherapy. Nature immunology. 2002;3(7):611–618. doi: 10.1038/ni0702-611. [DOI] [PubMed] [Google Scholar]
- 39.Leach DR, Krummel MF, Allison JP. Enhancement of antitumor immunity by CTLA-4 blockade [see comments] Science. 1996;271(5256):1734–1736. doi: 10.1126/science.271.5256.1734. [DOI] [PubMed] [Google Scholar]
- 40.Pandiyan P, Hegel JK, Krueger M, Quandt D, Brunner-Weinzierl MC. High IFN-gamma production of individual CD8 T lymphocytes is controlled by CD152 (CTLA-4) J Immunol. 2007;178(4):2132–2140. doi: 10.4049/jimmunol.178.4.2132. [DOI] [PubMed] [Google Scholar]
- 41.Fleissner D, Frede A, Knott M, Knuschke T, Geffers R, Hansen W, et al. Generation and function of immunosuppressive human and murine CD8+ T cells by transforming growth factor-beta and retinoic acid. Immunology. 2011;134(1):82–92. doi: 10.1111/j.1365-2567.2011.03469.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lenschow DJ, Walunas TL, Bluestone JA. CD28/B7 system of T cell costimulation. Annual review of immunology. 1996;14:233–258. doi: 10.1146/annurev.immunol.14.1.233. [DOI] [PubMed] [Google Scholar]
- 43.Walunas TL, Lenschow DJ, Bakker CY, Linsley PS, Freeman GJ, Green JM, et al. CTLA-4 can function as a negative regulator of T cell activation. Immunity. 1994;1(5):405–413. doi: 10.1016/1074-7613(94)90071-x. [DOI] [PubMed] [Google Scholar]
- 44.Gattinoni L, Ranganathan A, Surman DR, Palmer DC, Antony PA, Theoret MR, et al. CTLA-4 dysregulation of self/tumor-reactive CD8+ T-cell function is CD4+ T-cell dependent. Blood. 2006;108(12):3818–3823. doi: 10.1182/blood-2006-07-034066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pedicord VA, Montalvo W, Leiner IM, Allison JP. Single dose of anti-CTLA-4 enhances CD8+ T-cell memory formation, function, and maintenance. Proceedings of the National Academy of Sciences of the United States of America. 2011;108(1):266–271. doi: 10.1073/pnas.1016791108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Krummel MF, Allison JP. CTLA-4 engagement inhibits IL-2 accumulation and cell cycle progression upon activation of resting T cells. J Exp Med. 1996;183(6):2533–2540. doi: 10.1084/jem.183.6.2533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Williams MA, Tyznik AJ, Bevan MJ. Interleukin-2 signals during priming are required for secondary expansion of CD8+ memory T cells. Nature. 2006;441(7095):890–893. doi: 10.1038/nature04790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ayukawa H, Matsubara T, Kaneko M, Hasegawa M, Ichiyama T, Furukawa S. Expression of CTLA-4 (CD152) in peripheral blood T cells of children with influenza virus infection including encephalopathy in comparison with respiratory syncytial virus infection. Clinical and experimental immunology. 2004;137(1):151–155. doi: 10.1111/j.1365-2249.2004.02502.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jago CB, Yates J, Camara NO, Lechler RI, Lombardi G. Differential expression of CTLA-4 among T cell subsets. Clinical and experimental immunology. 2004;136(3):463–471. doi: 10.1111/j.1365-2249.2004.02478.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Brunet JF, Denizot F, Luciani MF, Roux-Dosseto M, Suzan M, Mattei MG, et al. A new member of the immunoglobulin superfamily--CTLA-4. Nature. 1987;328(6127):267–270. doi: 10.1038/328267a0. [DOI] [PubMed] [Google Scholar]
- 51.Leung-Theung-Long S, Mondor I, Guiraud M, Lamare C, Nagaleekar V, Paulet PE, et al. Impaired NFAT transcriptional activity in antigen-stimulated CD8 T cells linked to defective phosphorylation of NFAT transactivation domain. J Immunol. 2009;182(11):6807–6814. doi: 10.4049/jimmunol.0803539. [DOI] [PubMed] [Google Scholar]
- 52.Yang JC, Hughes M, Kammula U, Royal R, Sherry RM, Topalian SL, et al. Ipilimumab (anti-CTLA4 antibody) causes regression of metastatic renal cell cancer associated with enteritis and hypophysitis. J Immunother. 2007;30(8):825–830. doi: 10.1097/CJI.0b013e318156e47e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Royal RE, Levy C, Turner K, Mathur A, Hughes M, Kammula US, et al. Phase 2 trial of single agent Ipilimumab (anti-CTLA-4) for locally advanced or metastatic pancreatic adenocarcinoma. J Immunother. 2010;33(8):828–833. doi: 10.1097/CJI.0b013e3181eec14c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Prieto PA, Yang JC, Sherry RM, Hughes MS, Kammula US, White DE, et al. CTLA-4 Blockade with Ipilimumab: Long-Term Follow-up of 177 Patients with Metastatic Melanoma. Clin Cancer Res. 2012 doi: 10.1158/1078-0432.CCR-11-1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hodi FS, O'Day SJ, McDermott DF, Weber RW, Sosman JA, Haanen JB, et al. Improved survival with ipilimumab in patients with metastatic melanoma. The New England journal of medicine. 2010;363(8):711–723. doi: 10.1056/NEJMoa1003466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yuan J, Ginsberg B, Page D, Li Y, Rasalan T, Gallardo H, et al. CTLA-4 blockade increases antigen-specific CD8<sup>+</sup> T cells in prevaccinated patients with melanoma: three cases. Cancer Immunology, Immunotherapy. 2011;60(8):1137–1146. doi: 10.1007/s00262-011-1011-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Weber JS, Hamid O, Chasalow SD, Wu DY, Parker SM, Galbraith S, et al. Ipilimumab increases activated T cells and enhances humoral immunity in patients with advanced melanoma. J Immunother. 2012;35(1):89–97. doi: 10.1097/CJI.0b013e31823aa41c. [DOI] [PubMed] [Google Scholar]
- 58.Khattri R, Auger JA, Griffin MD, Sharpe AH, Bluestone JA. Lymphoproliferative Disorder in CTLA-4 Knockout Mice Is Characterized by CD28-Regulated Activation of Th2 Responses. The Journal of Immunology. 1999;162(10):5784–5791. [PubMed] [Google Scholar]
- 59.Lindsten T, Lee KP, Harris ES, Petryniak B, Craighead N, Reynolds PJ, et al. Characterization of CTLA-4 structure and expression on human T cells. The Journal of Immunology. 1993;151(7):3489–3499. [PubMed] [Google Scholar]
- 60.Freeman GJ, Lombard DB, Gimmi CD, Brod SA, Lee K, Laning JC, et al. CTLA-4 and CD28 mRNA are coexpressed in most T cells after activation. Expression of CTLA-4 and CD28 mRNA does not correlate with the pattern of lymphokine production. The Journal of Immunology. 1992;149(12):3795–3801. [PubMed] [Google Scholar]
- 61.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 62.Weinmann AS, Bartley SM, Zhang T, Zhang MQ, Farnham PJ. Use of chromatin immunoprecipitation to clone novel E2F target promoters. Molecular and cellular biology. 2001;21(20):6820–6832. doi: 10.1128/MCB.21.20.6820-6832.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]


