Figure 3. Global architecture of M3+TE.
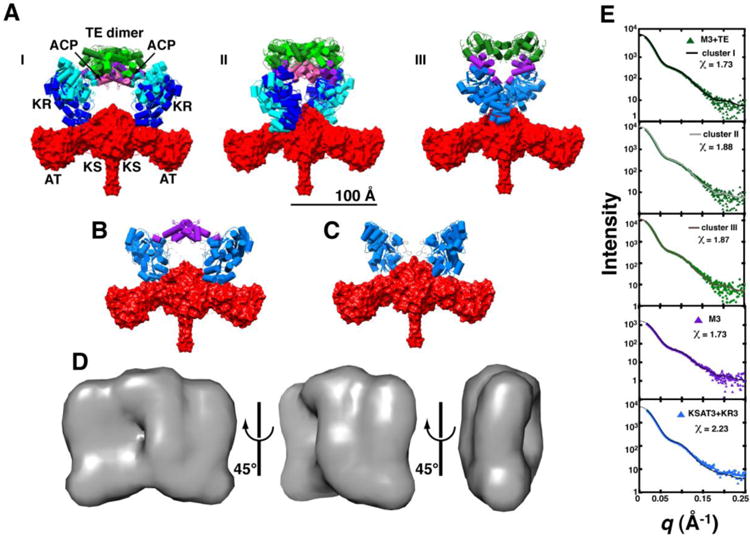
Ten independent rigid body refinement models were generated using CORAL with P2 symmetry applied and dimerization enforced across the KS and TE domains. The results were clustered using all-atom RMSD alignments under the default settings in DAMCLUST. (A) Cluster I contained 7/10 models (RMSD = 20 ± 6 Å), whereas 2/10 models were binned into cluster II (RMSD = 11.7 Å); the remaining structure is shown in panel III. Structures of each cluster were aligned using the KSAT3 didomain in order to assess heterogeneity at the KR, ACP, at TE domain positions. Two representatives from clusters I and II are shown. Domains are colored as in Figure 2B with dark and light shading indicating different cluster representatives. SAXS datasets were also collected for (B) M3 and (C) KSAT3+KR3, and processed in the same manner. Structures that closely resemble clusters I and II of M3+TE are shown. (D) Twenty independent ab initio calculations of the molecular envelope of M3+TE were performed using DAMMIF, with P2 symmetry imposed. The models were aligned, averaged, and filtered in DAMAVER to eliminate noise. The resulting shape is shown in three orientations. All structures are scaled equivalently with a 100 Å scale bar provided in (A). (E) Theoretical scattering curves for models generated via rigid body refinement were fit to the experimental datasets using CRYSOL. The resulting curves fit the data best in the low-q region (q < 0.15), suggesting a resolution of ∼40 Å (d = 2π/q). Chi2-free values are reported in Table S4.
