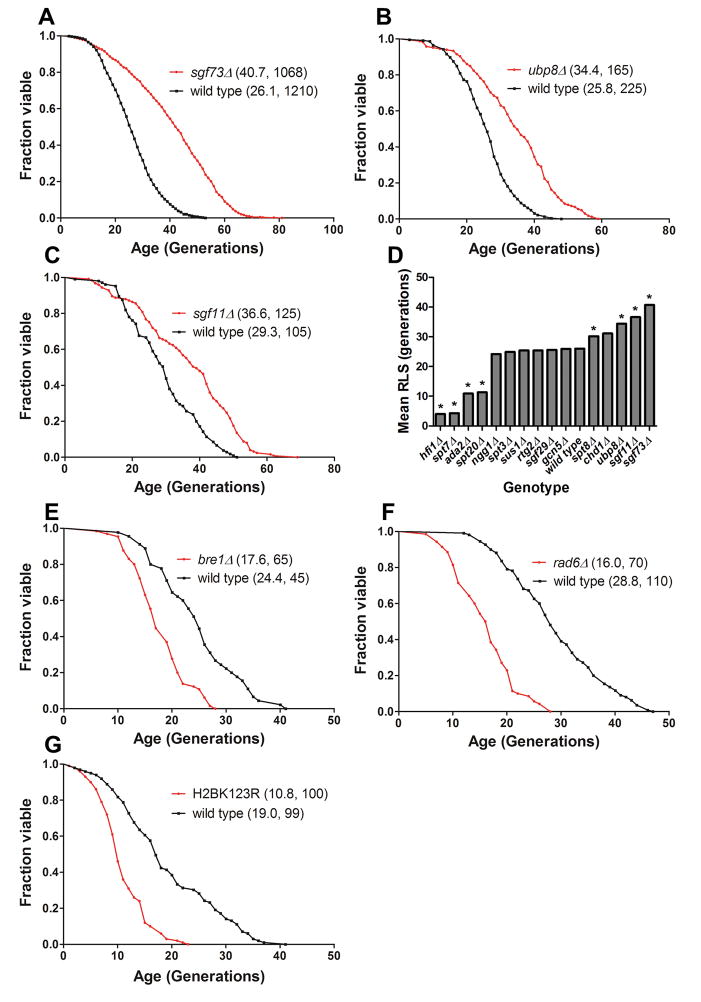
Figure 1.
Deletion of SAGA DUBm components SGF73, UBP8, and SGF11 significantly increases yeast RLS, and mutation of H2B-K123 or deletion of its monoubiquitinating enzymes shortens RLS. (A) sgf73Δ; (B)ubp8Δ ; (C) sgf11Δ; (D) summary of remaining viable SAGA component deletions; (E) bre1Δ; (F) rad6Δ; (G) H2B-K123R. Legends show (mean RLS, number of mother cells scored). See also Supplemental Figure S1, S2, and S3.