Abstract
Background and Aims Plant stature and shape are largely determined by cell elongation, a process that is strongly controlled at the level of the cell wall. This is associated with the presence of many cell wall proteins implicated in the elongation process. Several proteins and enzyme families have been suggested to be involved in the controlled weakening of the cell wall, and these include xyloglucan endotransglucosylases/hydrolases (XTHs), yieldins, lipid transfer proteins and expansins. Although expansins have been the subject of much research, the role and involvement of expansin-like genes/proteins remain mostly unclear. This study investigates the expression and function of AtEXLA2 (At4g38400), a member of the expansin-like A (EXLA) family in arabidposis, and considers its possible role in cell wall metabolism and growth.
Methods Transgenic plants of Arabidopsis thaliana were grown, and lines over-expressing AtEXLA2 were identified. Plants were grown in the dark, on media containing growth hormones or precursors, or were gravistimulated. Hypocotyls were studied using transmission electron microscopy and extensiometry. Histochemical GUS (β-glucuronidase) stainings were performed.
Key Results AtEXLA2 is one of the three EXLA members in arabidopsis. The protein lacks the typical domain responsible for expansin activity, but contains a presumed cellulose-interacting domain. Using promoter::GUS lines, the expression of AtEXLA2 was seen in germinating seedlings, hypocotyls, lateral root cap cells, columella cells and the central cylinder basally to the elongation zone of the root, and during different stages of lateral root development. Furthermore, promoter activity was detected in petioles, veins of leaves and filaments, and also in the peduncle of the flowers and in a zone just beneath the papillae. Over-expression of AtEXLA2 resulted in an increase of >10 % in the length of dark-grown hypocotyls and in slightly thicker walls in non-rapidly elongating etiolated hypocotyl cells. Biomechanical analysis by creep tests showed that AtEXLA2 over-expression may decrease the wall strength in arabidopsis hypocotyls.
Conclusions It is concluded that AtEXLA2 may function as a positive regulator of cell elongation in the dark-grown hypocotyl of arabidopsis by possible interference with cellulose metabolism, deposition or its organization.
Keywords: Arabidopsis thaliana, cell development, cell elongation, plant cell wall, expansins, expansin-like, extensiometry, hypocotyl growth, gravitropism
INTRODUCTION
Plant growth and shape result from carefully orchestrated cell divisions and mainly from the highly controlled and sometimes massive expansion of these newly formed cells. During expansion, the walls are centres of high activity. Primary walls of most dicotyledonous plants consist of xyloglucan (XyG)-tethered cellulose microfibrils embedded in a matrix of highly hydrated pectins and structural proteins (Carpita and Gibeaut, 1993). As plant cell walls can be considered as rather rigid boxes encapsulating the protoplast, their controlled weakening is crucial to allow turgor-driven expansion. It is well established that a wide variety of enzymes and factors influence and regulate the wall’s yielding properties, allowing an accurate control of the expansion process (Cosgrove, 2005). These include peroxidases (Passardi et al., 2006), β(1→4)-glucanases (Labrador and Nevins, 1989), xyloglucan endotransglucosylase/hydrolases (XTHs; Nishitani and Vissenberg, 2007), expansins (McQueen-Mason et al., 1992), yieldins (Okamoto-Nakazato et al., 2001) and lipid transfer proteins (LTPs; Nieuwland et al., 2005). For several of these enzyme families, enzymatic activities have been determined in vitro, but for yieldins and LTPs the mode of action still remains speculative.
Expansins were first discovered in studies of acid-stimulated growth as proteinaceous factors that have the capacity to induce extensibility and stress relaxation in cucumber hypocotyl walls in a pH-dependent manner (McQueen-Mason et al., 1992). This necessity for an acidic cell wall environment coupled to the cell wall-loosening properties links the acid growth theory, auxin and expansins (e.g. reviewed in Rayle and Cleland, 1992). Expansins show specific binding to cellulose, with a significant increase in binding strength when the cellulose is coated with hemicelluloses, suggesting possible interactions of expansins with XyG. Further experiments showed, however, that expansins do not possess hydrolytic activity and that they do not change the mass distribution of cucumber xyloglucans. These data led to the currently accepted model for expansin action: in vitro cell wall extension and in vivo cell expansion involve the disruption of the hydrogen bonds between cellulose microfibrils and tethering glucans in the wall by the expansins (McQueen-Mason and Cosgrove, 1994; Cosgrove, 2000a, b; Li et al., 2003; Sampedro and Cosgrove, 2005). This loosening would allow slippage of the cell wall polymers (creep) during turgor-driven cell wall extension, but prevents an irreversible weakening of the cell wall, which could result in detrimental lysis.
Expansins are classified into α- and β-expansins based on their sequence (Shcherban et al., 1995; Li et al., 2003; Choi et al., 2006). Expansins are small proteins (250–275 amino acids) composed of two compact domains [domain 1 (D1) and domain 2 (D2)] preceded by a signal peptide. Domain 1 is the N-terminal domain, homologous to the catalytic domain of proteins in the glycoside hydrolase family 45 (GH45). The C-terminal domain 2 is homologous to group-2 grass pollen allergens and has a sandwich fold. This is the cellulose-binding domain (CBD) (Cosgrove, 2000b, c; Sampedro and Cosgrove, 2005; Kerff et al., 2008). According to the recent nomenclature (Kende et al., 2004), only proteins with homology to both expansin domains should be designated expansins.
The effect of expansins on cell walls in vitro suggests the well-known cell wall-loosening function, yet roles in several other processes are also described. These include leaf abscission (Belfield et al., 2005), plant root colonization by Trichoderma reesei (Brotman et al., 2008), fruit ripening (Civello et al., 1999; Brummell et al., 2002; Fonseca et al., 2005), waterlogging-induced elongation (Cho and Kende, 1997; Caturla et al., 2002), cotton fibre elongation (Orford and Timmis, 1998), increasing cell wall flexibility upon cell wall folding during de- and rehydration in Craterostigma plantagineum (Jones and McQueen-Mason, 2004), root hair initiation (Cho and Cosgrove, 2002), pollen tube penetration (Cosgrove et al., 1997), hypocotyl and coleoptile elongation (Rochange et al., 2001; Gao et al., 2008), root elongation (Lee et al., 2003) and leaf development (Sloan et al., 2009).
Further attempts were made to better understand the working mechanism of expansins. Recently, the idea that expansins would bind not only to cellulose but also to a wider range of cell wall polymers was suggested (Nardi et al., 2013). By using a putative carbohydrate-binding module (CBM) from strawberry expansin 2 (CBM-FaExp2), the capacity to bind to xylan and pectin with different affinities was demonstrated. By this binding, the CBM-FaExp2 interfered with the activity of other cell wall-degrading enzymes, such as polygalacturonase, endo-glucanase, pectinase and xylanase (Nardi et al., 2013). Recently Goh and co-workers (2014) suggested that the specific effects of ectopic expansin expression depend on the degree of induction and the endogenous developmental pattern of organ growth. Transgenic arabidopsis plants with inducible cucumber expansin expression showed that arabidopsis was very sensitive to higher levels of expansin gene induction, even resulting in growth suspension, while the small increase in expansin expression led to an increase in lamina and petiole growth (Goh et al., 2014). These authors postulated that expansin only unlocks the molecular structure of the cell wall and that the observed growth depends on other apoplastic factors (Caderas et al., 2000; Rochange et al., 2001; Sloan et al., 2009).
It is striking that dicotyledonous plants with type I cell walls, such as Arabidopsis thaliana, also contain a minority of β-expansins besides α-expansins. These β-expansins are thought to bind to type II cell walls that are mainly found in monocots and grasses. For arabidopsis, 26 α-expansins and six β-expansins are described; for the monocot rice these numbers are 33 and 19, respectively (http://www.bio.psu.edu/expansins/). Blasting sequenced plant genomes with the known α- and β-expansin sequences revealed the additional presence of two families of expansin-like genes, which are significantly similar to the already known expansins. In arabidopsis, three expansin-like A (EXLA) members and one expansin-like B (EXLB) member were found (Sampedro and Cosgrove, 2005). Both expansin domains are present in EXLA and EXLB, but their amino acid sequences are different from those of α- and β-expansins (Li et al., 2002, 2003).
Despite the vast number of publications on α- and β-expansins, the exact functions of EXLA and EXLB in A. thaliana remain largely unclear (Choi et al., 2008; Dal Santo et al., 2013). However, different expansin-like proteins isolated from fungi or bacteria, such as swollenin expansin-like proteins from Trichoderma asperellum (Wang et al., 2011) or a cerato-platanin (CP) protein from the fungus Ceratocystis platani, are of interest to different research groups due to their cellulose biomass bioconversion abilities. CP has expansin-like activity and weakens filter paper in a concentration-dependent manner, and it represents a novel one-domain expansin-like protein with a hypothesized function in fungal interaction with the plant (Baccelli et al., 2014). Another example is bacterial Hc-EXLX2 from Hahella chejuensis (Lee et al., 2010), which enhanced the activity of cellulase and thereby increased the accessibility of other cell wall-degrading enzymes (Lee et al., 2010).
Here we investigate the expression and function of AtEXLA2 (At4g38400), a member of the EXLA family in A. thaliana, and discuss its possible role in cell wall metabolism and growth.
MATERIALS AND METHODS
Constructs
To generate the promoter::reporter constructs, a fragment 1044 bp upstream of the translation initiation site of At4g38400 was amplified by PCR from genomic DNA that was isolated as described by Edwards et al. (1991). Primers used for amplifying the promoter region were promEXLA2For (5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTTCTCGTCGACCAATGGATTGTCGGTG-3′) and promEXLA2Rev (5′-GGGGACCACTTTGTACAAGAAAGCTGGGTTTTTCTTGAAATTGAAGAATAACTAAAAAAAC-3′). The resulting product was gel-purified using the Ultra-Sep Gel Extraction Kit (Omega Bio-Tek), subsequently recombined with pDONR207 (Invitrogen) using GATEWAY™ technology (Invitrogen) and transformed into CaCl2-competent Escherichia coli MC1061 (Hoffman-La Roche Ltd). ENTRY-clones were screened by colony PCR, and selected clones were verified by bidirectional dideoxy-sequencing (VIB-Flemish Institute of Biotechnology) using primers AttL1 (5′-TCGCGTTAACGCTAGCATGGATCTC-3′) and AttL2 (5′-GTAACATCAGAGATTTTGAGACAC-3′). Sequence-verified ENTRY clones were recombined with destination vectors pGWB3 and pGWB4 (Nakagawa et al., 2007), leading to insertion of the promoter region upstream of the uidA or GFP sequence, respectively. After transformation in E. coli and colony PCR using primers promEXLA2for, GUSrev (5′-ATTGCCCGGCTTTCTTATA-3′) and GFPrev (5′-CCCTCGCCGGACAGCTGAA-3′), the final constructs were purified from E. coli using the Nucleospin® Plasmid QuickPure kit (Macherey-Nagel, Germany). The final constructs were then electroporated into Agrobacterium tumefaciens (strain C58 RifR harbouring pMP90; Koncz and Schell, 1986). Transformants were screened by colony PCR using the primers mentioned above and used for flower dip transformation of wild-type A. thaliana (Col-0, N1093) plants as described by Clough and Bent (1998) using a transformation buffer containing 5 % sucrose, MgCl2·6H2O (4 mm) and 0·02 % (v/v) Silwet L-77 (polyalkyleneoxide-modified heptomethyltrisiloxane) (GE Specialty Materials, Switzerland).
For the over-expression construct, the AtEXLA2 coding region was amplified from the cDNA clone RAFL04-09-M02 (pda00182) (Seki et al., 1998, 2002; Sakurai et al., 2005) using primers 35SEXLA2For (5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTTCATGCTGCAAGGCTTTCTCTTCCTC-3′) and 35SEXLA2Rev (5′-GGGGACCACTTTGTACAAGAAAGCTGGGTTTCAGTTCCAGATGTGATCGTCGC-3′). The product was recombined with pDONR207, and colonies were screened as described above. Correct ENTRY clones were recombined with pH2GW7·0 (Karimi et al., 2002, 2007) and screened by colony PCR using primers 35SEXLA2For and OVrev (5′-GCTCAACACATGAGCGAAAC-3′). Transformation of agrobacterium and arabidopsis was as described above, except that the corresponding primers were used.
Plant growth conditions
Wild-type (Col-0) and mutant seeds (SALK_147678) of A. thaliana were obtained from the Nottingham Arabidopsis Stock Centre (Scholl et al., 2000). For plant growth on plates, seeds were surface sterilized for 5 min in 6 % commercial bleach followed by three rinses in sterile water before they were placed in rows on half-strength MS Petri plates containing 2·2 g L–1 Murashige and Skoog salts including vitamins (Duchefa, The Netherlands), 1 % (w/v) sucrose (Duchefa), pH 5·7 (KOH) and 8 g L–1 plant agar (Duchefa). Plates were subsequently stratified for 4 d at 4 °C in the dark. Plates were then placed in a growth chamber (1·5E + 15 photons cm–2 s–1; 16 h light/8 h dark; 24 °C). Images of plates were taken at the time points as mentioned in the Results, using a digital camera (Canon, 50D), and measurements were performed using ImageJ (Abramoff et al., 2004).
For measurements of dark-grown hypocotyls, seeds were sown on solid ES medium (Estelle and Sommerville, 1987) and stratified for 3 d at 4 °C. The synchronous germination was induced by exposure to fluorescent white light (150 μmol m–2 s–1 True Light; Philips, Eindhoven, The Netherlands) for 4 h at 21 °C. The transfer to light is referred to as ‘induction’ and considered as time zero for all the experiments. Darkness was obtained by wrapping the Petri dishes in four layers of aluminium foil. Covered plates were afterwards placed vertically at 21 °C in an environmentally controlled growth cabinet (cooled incubator BRC120, Bioconcept-Firlabo, Beun De Ronde, Belgium). Images of plates were taken at the time points as mentioned in the Results under green safe light to avoid any white light inhibition, using a digital camera (Canon, 50D). Manipulation of seedlings had no effect on hypocotyl growth or final length. Hypocotyl length was measured as the distance between the top of the root hairs around the collet, and the base of ‘V’ made by the petioles of the cotyledons (Scheres et al., 1994). The length of the hypocotyls was measured using ImageJ software (Abramoff et al., 2004).
For experiments on medium containing hormones or precursors, stocks of the studied compounds were made in dimethylsulphoxide (DMSO) and added to the medium when it had reached a temperature of <50 °C after autoclaving.
For the gravitropism experiment, half-strength MS medium (without sucrose) was used. Seeds were prepared as mentioned above and, after 3 d of stratification at 4 °C, plates were placed vertically in the growth chamber (1·5 × 1015 photons cm–2 s–1; 16 h light/8 h dark; 24 °C). Subsequently, after 3 d of growing, plates were turned 90 ° (Ishikawa and Evans, 1997; Hou et al., 2004; Young et al., 2006; Miller et al., 2007). This moment of turning is considered as time zero for this experiment. Plates with seedlings were collected every hour till 10 h after the turning and stained in β-gluronidase (GUS) solution on the plates in order to keep the real position of the root. Images of seedlings were taken under bright field illumination using a Zeiss Axioskope (Zeiss, Jena, Austria) equipped with a Nikon DXM 1200 or Nikon DS-Fi1 digital camera (Nikon Instruments Inc., Melville, NY, USA).
To prepare leaf series, seeds of the wild type and two over-expressing lines were sown in the soil. Seeds were stratified for 3 d at 4 °C in order to obtain synchronous germination. Subsequently, trays with seeds were placed in a growth chamber (1·5E + 15 photons cm–2 s–1; 16 h light/8 h dark; 24 °C) and plants were collected at different developmental stages (14, 20, 26 and 32 d old). Cotyledons and leaves were dissected from the stem under a binocular microscope (Nikon SMZ-1B). Images of leaf series were taken using a Nikon AZ100 Multizoom equipped with a Nikon DS-Ri1 digital camera and in DIC (differential interference contrast) mode. The area and roundness of the leaves and length of the petioles were measured using ImageJ (Abramoff et al., 2004).
Identification of over-expression lines
Homozygous plants were grown for 2 weeks on half-strength MS medium before harvesting the plant material. Total RNA was extracted from leaves and roots using the Concert™ Plant RNA reagent (Invitrogen) and concentrations were determined on a nanodrop (model ND1000, Montchanin, DE, USA). A 1 μg aliquot of RNA was subsequently treated with RQ1 RNase-free DNase I (Promega, USA) according to the manufacturer’s instructions in a total volume of 10 μL at 37 °C for 27 min and heat-inactivated at 65 °C for 15 min. This reaction was subsequently used for cDNA synthesis with a mixture of random hexamers and oligod(T)18 primers using Superscript II reverse transcriptase (Invitrogen). For qPCR analysis, a capillary-based LightCycler (Roche Diagnostics, Belgium) was used in combination with SYBR green containing Maxima™ SYBR Green Master Mix (Fermentas, catalogue no. K0252) (ROX was omitted). As template 12·5 ng (5 μL of 1/20 diluted cDNA) was used in a reaction of 25 μL. Before quantification, the efficiency of primers was determined over six concentrations ranging from 0 to 50 ng. Expression ratios were calculated according to Pfaffl (2001). Primers used to amplify reference genes were Forwβ-actRT (5′-GCGATGAAGCTCAATCCAAA-3′), Revβ-actRT (5′-GTCACGACCAGCAAGATCAAG-3′), ForwEF-1αRT (5′-ATGCCCCAGGACATCGTGATTTCAT-3′) and RevEF-1αRT (5′-ACCTAGCCTTGGAGTATTTGGGGG-3′). AtEXLA2 transcripts were detected using the primers FEXLA2RT (5′-CTTGTCCTTAGCAGCAGAGCC-3′) and REXLA2RT (5′-GGTACAAGAGCTTTATCGCC-3′).
GUS staining
Histochemical GUS stainings were performed according to a modified protocol of Jefferson et al. (1987). Plant tissues were submerged in a staining solution [potassium phosphate buffer 200 mm (pH 7·0), potassium ferrocyanide 1 mm, potassium ferricyanide 1 mm, disodium EDTA 10 mm, 5-bromo-4-chloro-3-indolyl-β-d-glucuronide 1 mm] and incubated overnight at 37 °C. To improve the penetration of the staining solution in tissues containing a cuticle, samples were briefly incubated in 70 % acetone prior to staining. After staining, tissues were rinsed 3 × 10 min in H2O, subsequently fixed in ethanol:acetic acid (3:1, v/v) and cleared in 8 m NaOH. Images were taken under bright field illumination using a Zeiss Axioskope equipped with a Nikon DXM 1200 or Nikon DS-Fi1 digital camera (Nikon Instruments Inc.). Representative images of at least three independent transgenic lines are shown.
Transmission electron microscopy
Seedlings of 35S::AtEXLA2 6_2 and wild-type Col-0 were grown for 4 d in the dark as described above before being prepared as described by Roland et al. (1982). The hypocotyls were first cut into three pieces (upper, middle and basal sections) and the sections were marked with an aqueous solution of ruthenium red so that the orientation would be known during embedding. Subsequently samples were fixed for 2 h in 3 % glutaraldehyde in 0·1 m Na-cacodylate buffer (pH 7·4) and washed three times for 10 min in the same buffer without fixative. At 1 h post-fixation with 1 % OsO4 in the same buffer, they were rinsed three times for 10 min in the buffer only. Hypocotyl sections were extracted for 10 h in 40 % methylamine under continuous mild stirring and rinsed three times for 10 min in distilled water. Samples were then dehydrated for 10–15 min in a graded ethanol series (30, 50, 70, 80, 95 % and twice in 100 %) and embedded in Spurr resin following the manufacturer’s instructions (SPURR: Modified SPURR Embedding Kit). Polymerization was performed by heating the samples at 70 °C for 8 h. Ultrathin transverse sections (90–100 nm) were made at the level of the apical hook, middle and the basal part of the hypocotyl. Observations were made with a Tecnai G2 SpiritBio Twin microscope equipped with a Veleta camera under a power of 120 kV.
Extensiometry
The biomechanics of hypocotyls was studied using the constant-load (creep) method. Etiolated 4-day-old wild-type and 35S::AtEXLA2 6_2 arabidopsis seedlings were transferred individually into 1·5 mL Eppendorf test tubes, frozen by plunging the closed tubes into liquid nitrogen, stored at −20 °C and used for extensiometry within 2 weeks after freezing. For heat inactivation, distilled water (1·5 mL, 22 °C) was added to Eppendorf test tubes with frozen sampes, and the tubes were transferred to a pre-heated (90 °C) thermomixer (Eppendorf Thermomixer® comfort) and incubated with shaking at 100 rpm for 3 min. Then, the heat inactivation was rapidly stopped by transferring the tubes to a 1000 mL beaker with water at 22 °C. Finally, the frozen–thawed heat-inactivated arabidopsis seedlings were transfered to small Petri dishes filled with distilled water and kept in a refrigerator at 4 °C for 1–2 h before being used in creep tests.
In vitro extension of frozen–thawed arabidopsis hypocotyls was measured with a custom-built constant-load extensiometer described in Suslov and Verbelen (2006) using the same procedure as in Miedes et al. (2013). A 5 mm long hypocotyl segment (starting from 1·5 mm below the cotyledons) was secured in the extensiometer and pre-incubated in a buffer (20 mm MES-KOH, pH 6·0 or 20 mm Na-acetate, pH 5·0) in the relaxed state for 2 min, after which it was extended in the same buffer under a constant load (400, 500, 600 or 700 mg) for 15 min. The creep rate was calculated with the formula given in Vandenbussche et al. (2011).
The cross-sectional area of the hypocotyl cell walls used in extensiometry was determined by measuring their dry weight per unit length (Cleland, 1967) as detailed in Miedes et al. (2013).
RESULTS
AtEXLA2 expression patterns
In order to investigate the potential functions of AtEXLA2, a detailed expression analysis was performed using the arabidopsis eFP browser (Winter et al., 2007; Supplementary Data Figs S1 and S2) and promoterAtEXLA2::reporter lines. Both uidA and GFP were used as reporters, but only GUS lines were analysed in detail.
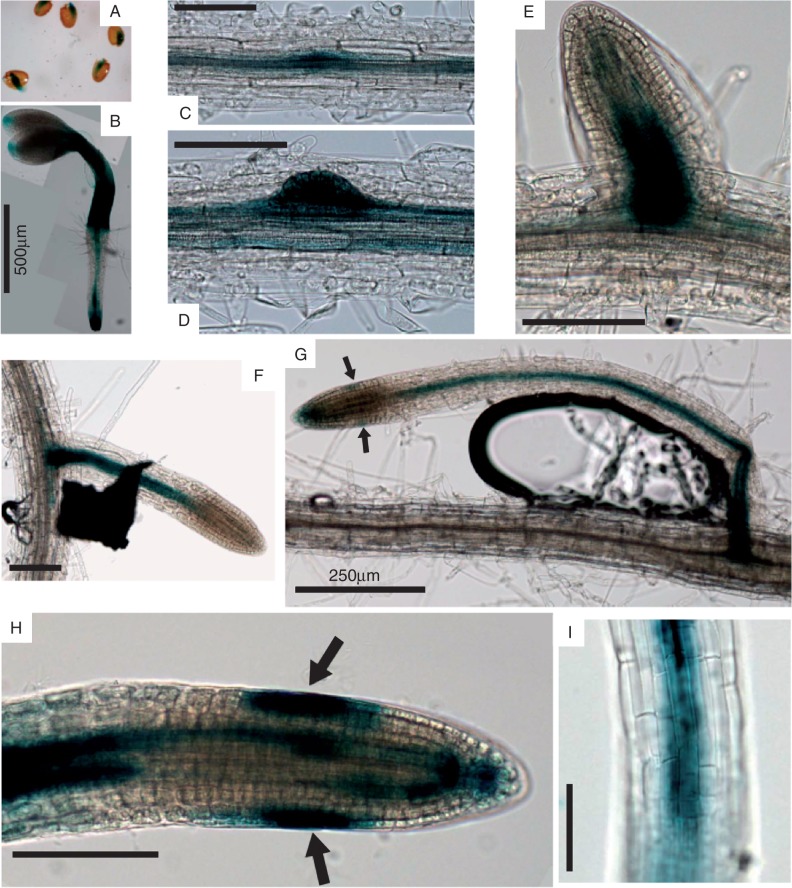
Staining with GUS was observed in seeds before and during testa rupture, suggesting a potential role for AtEXLA2 during embryonic growth or seed germination (Fig. 1A). In very young seedlings (2 d old), it was obvious that AtEXLA2 expression was present both in the hypocotyl and in the root. AtEXLA2 is strongly expressed throughout the hypocotyl, while weak expression is seen only in the tip of the cotyledon, presumably around the central hydathode. In young roots, expression was strongest in the root tip, with a maximum in the central cylinder starting at the onset of the elongation zone. In the differentiation zone of the root, expression was limited to the central cylinder (Fig. 1B). In roots of 8-day-old seedlings AtEXLA2 expression mimicked that of younger roots, with the addition of clear expression in all stages of lateral root primordia development (Fig. 1C–G). During the initial lateral primordia divisions, expression was present throughout the new primordia (Fig. 1C, D); in older stages AtEXLA2 expression was confined to the central cylinder of the new lateral root (Fig. 1E–G). Weak GUS staining in the central cylinder was present in the meristem from the quiescent centre on, and became much stronger as soon as cells entered the elongation zone (Fig. 1H, I). As primary and lateral roots matured, the oldest cells belonging to the lateral root cap (which also undergo a phase of cell elongation) and to the columella showed strong GUS staining (see arrow in Fig. 1G, H).
Fig. 1.
GUS staining in seedlings and roots of promoterAtEXLA2::uidA plants. (A) Seedlings emerging from seeds. (B) Two-day-old seedling. (C–G) Staining at different developmental stages of lateral root development. (G) Magnification of the lateral root tip with staining in the central cylinder, elongating lateral root cap cells (see arrows), columella and quiescent centre cells. (H) Primary root grown on ES medium showing the increase in expression in the central cylinder when cell elongation commences. Arrows indicate staining in elongating lateral root cells as in (G). (I) Close-up of the elongation zone with expression in the central cylinder. Scale bars represent 100 μm, unless indicated otherwise.
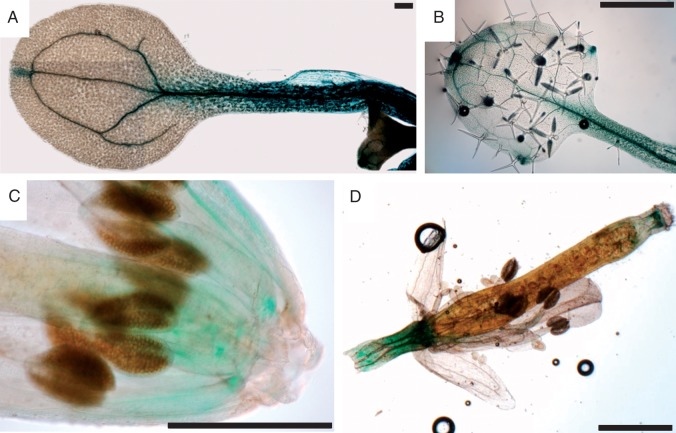
It was reported before that over-expression of expansins affects different aspects of plant growth (Choi et al., 2006); therefore, the expression of AtEXLA2 was also investigated in the above-ground parts of seedlings and adult plants. Cotyledons showed strong GUS staining throughout the whole petiole and in the vasculature (Fig. 2A). An additional site of GUS staining was localized at the central hydathode of the cotyledon (Fig. 2A), while the other cotyledon cells showed no AtEXLA2 expression at all. In older soil-grown plants, GUS staining was found in a pattern similar to that in the cotyledons. The first two leaves showed a similar pattern, with staining concentrated in or around the veins of the petiole and leaf blade and the most intense staining in the midvein of the leaf and the petiole (Fig. 2B). In the third and the fourth leaf, staining was not visible in the veins but was concentrated around the hydathodes, a pattern that was also present in the cauline leaves (results not shown). In flowers, staining was mainly present in the veins of filaments (Fig. 2C). Also the peduncle and the zone just underneath the papillae showed GUS staining (Fig. 2D).
Fig. 2.
GUS staining in cotyledons, leaves and inflorescences of promoterAtEXLA2::uidA plants. (A) Cotyledon, (B) young leaf, (C) inflorescence, (D) detail of stamen. Scale bars represent 1 mm.
Sensitivity of AtEXLA2 expression for auxin
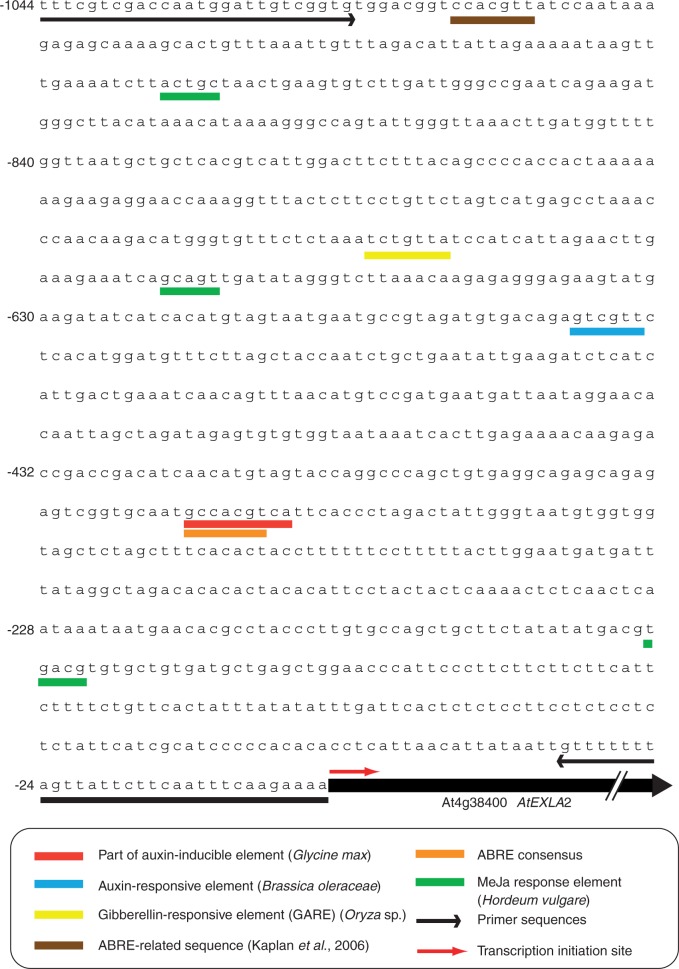
As expansins are accepted to be involved in acid growth and since cell wall acidification by plasma membrane-localized H+-ATPases can be induced by auxin, we checked whether the AtEXLA2 promoter is auxin responsive. We performed an in silico analysis of the promoter sequence (Fig. 3), revealing sequence similarities to a part of an auxin-inducible element of Glycine max and of an auxin-responsive element of Brassica oleraceae, but not to auxin-specific cis-elements. In addition, some potential cis-elements related to abscisic acid (ABA) and methyl jasmonate were detected. A further analysis using Genevestigator (Zimmermann et al., 2004) indicated that the expression of At4g38400 changes after auxin and ABA application. To check the validity of the in silico results, the effects of different compounds on the GUS expression pattern were studied. No visible differences in GUS staining patterns were found when either indole acetic acid (IAA), ABA, naphthalene acetic acid (NAA), 1-naphthylphthalamic acid (NPA), 1-aminocylcopropane-1-carboxylicacid (ACC), l-α-(2-aminoethoxyvinyl)glycine (AVG), GA3 or jasmonic acid was added to seedlings (results not shown).
Fig. 3.
In silico analysis of the promoter region of At4g38400. In silico promoter analysis of AtEXLA2 (At4g38400) using PlantCARE (Rombauts et al., 1999; Lescot et al., 2002) and PLACE (Higo et al., 1999). The sequences underlined with arrows represent the primers used for PCR amplification of the promoter. Cis-elements are colour coded.
Comparison of AtEXLA2 gene structure with AtEXPA, AtEXPB, AtEXLA and AtEXLB family members
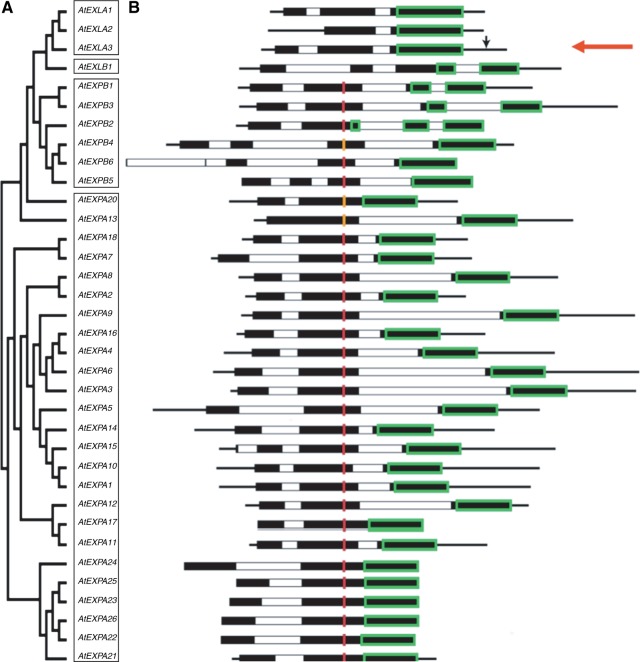
A graphical display of all the expansin and expansin-like genes of arabidopsis reveals that most AtEXPA genes have two introns, all AtEXPB genes have three introns, AtEXLA1 and 2 have two introns, AtEXLA3 has only one intron, and AtEXLB1 has three introns (Fig. 4). In this figure, those regions of the exons that code for the catalytic site of expansins (D1) and the CBD (D2) are indicated with red and green boxes, respectively. This analysis indicates that only the CBD is present in AtEXLA2 (see red arrow), while the catalytic site is clearly absent.
Fig. 4.
Phylogenetic and structural analysis of Arabidopsis thaliana expansins. (A) Phylogenetic tree of A. thaliana expansin proteins. The different expansin subfamilies are boxed. (B) Graphical representation of the gene structure of the corresponding expansins from (A). White and black bars depict introns and exons, respectively; black lines represent 3′- and 5′-untranslated regions. The regions coding for the presumed carbohydrate-binding modules are boxed in green, and the coding region for the presumed catalytic residues (HFD) are shown in red. Where the catalytical triad differed by only one amino acid, it is shown in orange. The T-DNA insertion in AtEXLA2 is indicated by a black arrow. Sequences are aligned to the catalytical triad where possible.
Effect of altered AtEXLA2 expression levels on development
To investigate the effects of lowered or absent AtEXLA2 expression levels on plant development, the only available T-DNA insertion line (SALK_147678) in AtEXLA2 was obtained and homozygous plants were selected by PCR screening. Unfortunately sequencing of a PCR product covering the T-DNA border indicated that the T-DNA insertion was localized 127 bp after the stop codon (see the vertical black arrow in Fig. 4). Semi-quantitative PCR of AtEXLA2 expression levels did not show visible differences in the Salk line compared with the wild type. The presence of the At4g38400 transcript at wild-type levels in the homozygous Salk line was confirmed with qPCR (results not shown).
To overcome the lack of a suitable knock-out plant and to avoid the potential problems with redundancy, plants over-expressing AtEXLA2 were generated. The AtEXLA2-coding region was placed downstream of the Cauliflower mosiac virus (CaMV) 35S promoter in pH2GW7, 0 (Karimi et al., 2002, 2007) and transformed to wild-type (Col-0) plants. Homozygous T3 plants harbouring a single insertion event were selected, and AtEXLA2 expression levels were determined in 7-day-old seedlings using qPCR. Two lines with high over-expression levels compared with the wild type (339 × in line_6_2 and 73 × in line_3_5, respectively) were selected for further experiments.
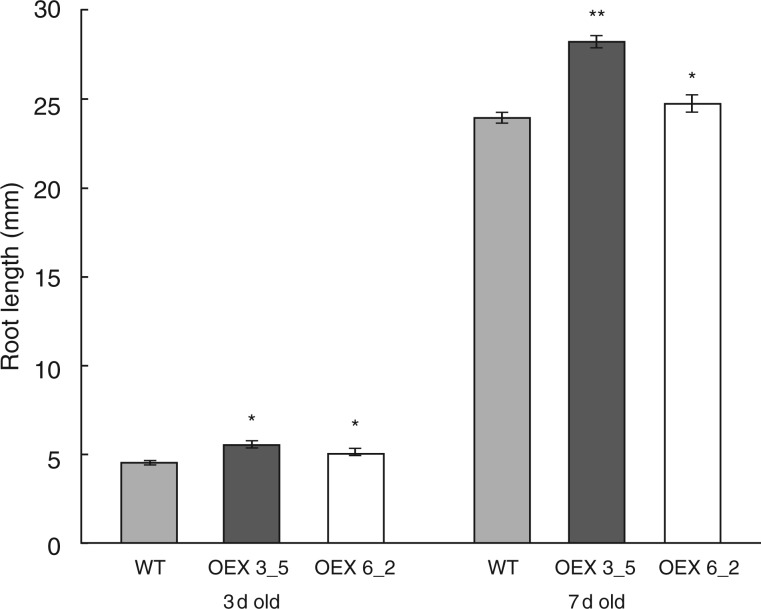
The strong expression of AtEXLA2 in the root (Fig. 1) suggested an involvement in root development. Therefore, the effect of AtEXLA2 over-expression on root growth was studied. After 3 and 7 d of growth, roots of the two over-expressing lines were significantly longer than those of the wild type (Fig. 5).
Fig. 5.
Root length in the wild type (WT) and two 35S::AtEXLA2 lines. The WT (Col-0) and two CaMV35S::AtEXLA2 lines (line 6_2 and 3_5) were grown in the light on half-strength MS medium for 3 and 7 d and their root length was scored (mean ± s.e.). Asterisks indicate significant differences.
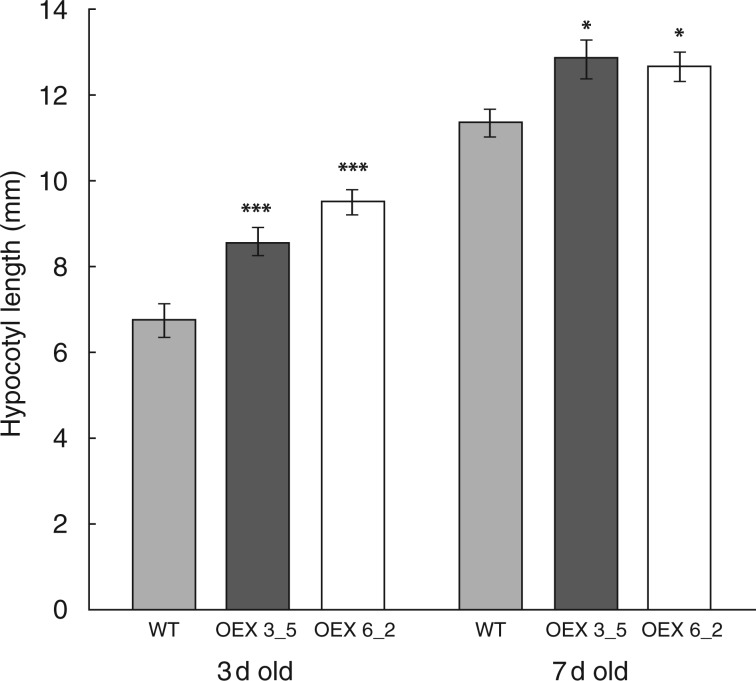
As well as being expressed in the root, GUS staining patterns suggested prominent expression of AtEXLA2 in the above-ground parts of the plant, including the hypocotyl and cotyledons. The length of 3- and 7-day-old dark-grown hypocotyls was determined. Under standard conditions, hypocotyls of over-expressing plants were significantly longer than in wild-type plants at both day 3 (142 and 128 % for lines 6_2 and 3_5, respectively) and day 7 (111 and 113 % for lines 6_2 and 3_5, respectively) [t-test, P < 0·01 (day 3) and P < 0·05 (day 7); Fig. 6]. The leaf area of cotyledons and real leaves (14, 20, 26 and 32 d old) was studied in the two over-expression lines, but did not show differences compared with the wild type (results not shown). Also no significant differences were found in vein pattern or petiole length (results not shown).
Fig. 6.
Dark-grown hypocotyl length in the wild type (WT) and two 35S::AtEXLA2 lines. The WT (Col-0) and two CaMV35S::AtEXLA2 lines (line 6_2 and 3_5) were grown in the dark on ES medium for 3 and 7 d and their hypocotyl length was measured (mean ± s.e.). Asterisks indicate significant differences.
Plant roots and stems bend in response to gravity in a process called gravitropism (Blancaflor, 2002; Miller et al., 2007). PromoterAtEXLA2::reporter lines have shown GUS staining in the elongation zone and lateral root cap cells (Fig. 1H, I), both zones being involved in the auxin-dependent gravitropic reaction. The expression pattern of AtEXLA2 genes was examined during gravitropism by turning the Petri plates 90 ° and checking the GUS signal every hour until 10 h after the turning. The GUS expression pattern did not change when compared with the normal growth situation (results not shown).
Effect of AtEXLA2 over-expression on cell wall thickness
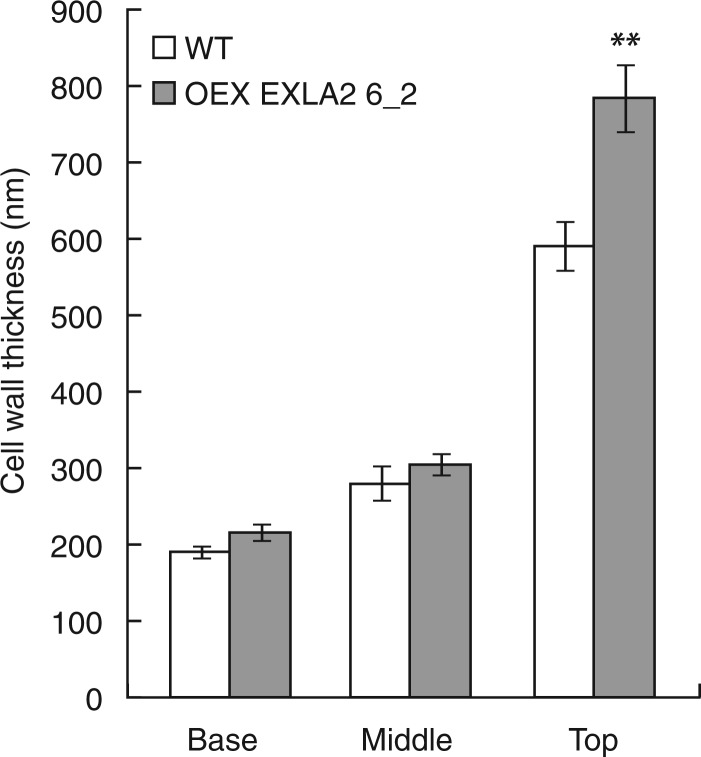
To study the effect of AtEXLA2 on cell wall thickness, 4-day-old etiolated hypocotyls were sampled and the thickness of the outer epidermal cell wall was measured at the bottom, middle and upper part of the hypocotyl. At this moment, cells at the base have undergone rapid elongation so that their walls are at their thinnest, cells in the middle part are undergoing fast elongation and cells in the upper part are still in the slow growth phase where a thick cell wall is deposited and just before the rapid elongation will start. From Fig. 7 it becomes clear that in the upper part, cell walls are thicker in over-expression line 6_2 than in the wild type, while they are equally thin in the other two zones.
Fig. 7.
Cell wall thickness in the wild type (WT) and 35S::AtEXLA2. Ultrathin sections were prepared at the base, middle and top of 4-day-old etiolated hypocotyls of the WT and 35S::AtEXLA2 line 6_2. Samples were studied with a transmission electron microscope and the thickness of the external epidermal cell wall was measured. Asterisks indicate significant differences.
Effect of AtEXLA2 over-expression on cell wall extensibility
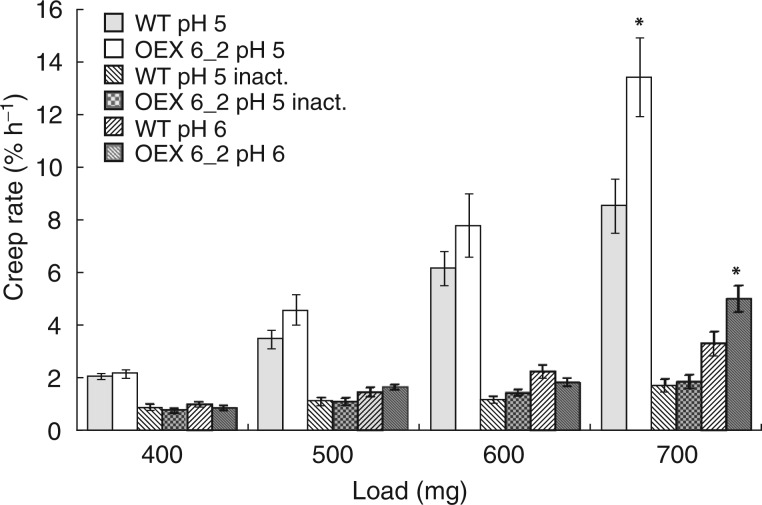
Cell wall extensibility plays a crucial role in the control of plant cell growth at the biophysical level. Thus the cell well extensibility was estimated in frozen–thawed wild-type and 35S::AtEXLA2 6_2 hypocotyls using the creep test (Fig. 8). Four-day-old growing hypocotyls were chosen, because they were sufficiently long for clamping in the extensiometer. At this stage, wild-type and 35S::AtEXLA2 6_2 hypocotyls had growth rates that did not differ significantly. In an attempt to predict the effect of different turgor values on the wall deformation, the hypocotyls were extended under a range of loads. The wall cross-sectional area in the 5 mm long sub-apical zone of hypocotyls extended in the creep test was similar in the wild-type and 35S::AtEXLA2 6_2 plants (results not shown) when estimated from the wall dry weight per unit length (Miedes et al., 2013). This zone includes the middle part of hypocotyls analysed in Fig. 7, so the results on the wall cross-sectional area obtained by the two alternative methods are consistent. Equal cell wall cross-sectional areas in the wild-type and 35S::AtEXLA2 6_2 plants indicate that each constant load generates a similar stress in their hypocotyl cell walls.
Fig. 8.
Creep rate of frozen–thawed arabidopsis hypocotyls. The creep rate of frozen–thawed wild type (WT) and 35S::AtEXLA2 6_2 (OEX 6_2) hypocotyls was measured under four different loads at pH 5 and pH 6 without heat inactivation, and pH 5 after heat inactivation (inact.) (mean ± s.e., n = 8–9). Asterisks indicate significant differences between the over-expression line and the WT under the same load and pH value. In all cases, the creep rate at pH 5 was significantly higher than that at pH 6 and pH 5 after heat inactivation (not shown on the plot).
The creep rates of hypocotyls were compared at pH 5, pH 6 and pH 5 with heat inactivation. A pH 5 buffer was used because expansins are activated at pH ≤5·5 (McQueen-Mason et al., 1992). They lose their activity at pH 6, but this pH value is optimal for different classes of cell wall-loosening proteins such as XTHs (Maris et al., 2011). The heat treatment was used to inactivate essentially all cell wall-loosening proteins to reveal their contribution to wall creep.
Hypocotyls of wild-type and 35S-AtEXLA2 6_2 plants demonstrated a strong acid-induced creep in a pH 5 buffer under all the loads studied (Fig. 8). This presumably expansin-mediated effect was completely eliminated by a heat treatment. The creep rate of heat-inactivated hypocotyls extended at pH 5 dropped to or below that found at pH 6 without heat inactivation. No significant differences were found in the creep rate of the wild type and the over-expression line under 400–600 mg loads, consistent with their in vivo growth rates. A marked proportion of hypocotyls broke under a 700 mg load (Table 1), and this was accompanied by a very different trend for in vitro cell wall extension. The creep rate of 35S::AtEXLA2 6_2 hypocotyls was significantly higher than in the wild type at both pH 5 and pH 6 under a 700 mg load (Fig. 8). Interestingly, hypocotyls of the over-expression line broke twice as often as wild-type hypocotyls under the maximal load used (Table 1). These results show that AtEXLA2 over-expression did not affect the ability of the wall to extend under moderate uniaxial stress values. On the other hand, the biomechanics under the maximal load suggests that the over-expression decreases the wall strength. The high creep rate of 35S::AtEXLA2 6_2 hypocotyls at pH 5 and pH 6 under a 700 mg load could precede their failure or just be an indicator that their ultimate strength is approaching. This burst increase in the creep of the over-expression line is not seen after a heat treatment, presumably because of failure of hypocotyls (Table 1) that would demonstrate a high creep rate without heat inactivation.
Table 1.
Frequency of hypocotyl failures in the creep test under different loads
| 400 mg | 500 mg | 600 mg | 700 mg | |
|---|---|---|---|---|
| WT pH 5 | 0·0 %* (8/8)† | 0·0 % (8/8) | 0·0 % (8/8) | 27·3 % (8/11) |
| OEX 6_2 pH 5 | 0·0 % (8/8) | 0·0 % (8/8) | 20·0 % (8/10) | 20·0 % (8/10) |
| WT pH 5 inactivated | 0·0 % (8/8) | 11·1 % (8/9) | 27·3 % (8/11) | 27·3 % (8/11) |
| OEX 6_2 pH 5 inactivated | 0·0 % (8/8) | 11·1 % (8/9) | 11·1 % (8/9) | 55·6 % (8/18) |
| WT pH 6 | 0·0 % (9/9) | 0·0 % (9/9) | 25·0 % (9/12) | 25·0 % (9/12) |
| OEX 6_2 pH 6 | 0·0 % (9/9) | 0·0 % (9/9) | 18·2 % (9/11) | 25·0 % (9/12) |
*The frequency of hypocotyl failures expressed as a percentage of total measurements.
†The first number in parentheses indicates the number of successful creep measurements without hypocotyl failures, and the second number after a slash refers to the total number of creep measurements.
OEX6_2 represents 35S::AtEXLA2 line 6_2.
DISCUSSION
Plant growth is the result of an increase in the number of cells and, more importantly, the volume of these newly formed cells through cell expansion. For the latter process, the initially deformation-resistant cell walls need to be modified. The ability of cell walls to undergo irreversible turgor-driven expansion is largely determined by the physical properties of the cell wall and by apoplastic enzymes modifying and/or restructuring its components (Cosgrove et al., 2000a, b, c; Thompson and Fry, 2001). For several protein families, the ability to induce cell wall extension in vitro or/and in vivo has been experimentally demonstrated (Labrador and Nevins, 1989; Okamoto-Nakazato et al., 2001; Nieuwland et al., 2005; Passardi et al., 2006; Van Sandt et al., 2007). In the same year as XTHs were discovered (Fry et al., 1992), the existence of another class of proteins with cell wall-loosening capacities, i.e. eexpansins, was proven (McQueen-Mason et al., 1992). Phylogenetic and sequence analyses have shown that expansin sequences are present in early diverging land plants, such as the liverwort Marchantia and the water fern Marsilea. These data point to a likely early evolutionary origin in the algal lineage (Cosgrove, 2000c). Moreover, expansin-coding sequences are even found in non-plant species. However, the gene structure from expansins of non-plant species such as a mycetozoan slime mould (Dictyostelium discoideum), fungi, mussels and a Solonaceous nematode (Globodera rostochiensis) (Xu et al., 2001; Li et al., 2002; Darley et al., 2003; Qin et al, 2004; Kudla, 2005; Brotman et al., 2008) differs from that in plants, suggesting that expansins have evolved separately in plant and non-plant species (Choi et al., 2006). Taken together with the vast number of expansins in some species (mostly due to gene duplication), this clearly indicates that expansins fulfil a crucial function during plant life.
Several characteristics are conserved in the expansin and expansin-like genes. First of all, the overall gene structure is conserved in expansin gene families. All expansins contain four conserved introns, except α-expansins, which lack the second and fourth intron. The signal peptide is also conserved in all expansin(-like) proteins. The N-terminal part of the protein contains the presumed catalytic domain that contains several conserved cysteines that are responsible for the secondary structure of the catalytic domain. In expansin-like A and B subfamilies, several of these conserved cysteines are not present, while expansin-like A proteins have two cysteines in their C-terminal domain. The catalytic site itself is believed to be represented by the amino acids HFD (histidine–phenyalanine–aspartate), and, strikingly, is absent in members of the expansin-like A and B groups (Fig. 4). This strongly suggests that expansin-like proteins cannot show classical expansin activity. However, this does not preclude the potential presence of an activity which differs from that described for α- and β-expansins. The C-terminal part of expansins has been described as a putative CBD, based on the similarity to the CBD of cellulase (Cosgrove, 2000c). From a similar analysis, it seems that AtEXLA2 (At4g38400) only contains the CBD and not the catalytic site that is typical for α- and β-expansins.
The in silico expression pattern analysis of the At4g38400 gene using the arabidopsis eFP browser (Winter et al., 2007; Supplementary Data Figs S1 and S2), suggests enriched expression in roots, hypocotyls and leaves, and this was fully confirmed in independent promoterAtEXLA2::reporter lines, indicating that the cloned promoter region was representative. In the root, AtEXLA2 expression was strongest in the elongation zone, quiescent centre, columella cells and stele, while in older roots clear expression was observed at all stages of lateral root primordia development. The expression analysis clearly shows that AtEXLA2 expression is detected in elongating cells of the root, hypocotyl and petioles, which suggests the involvement of AtEXLA2 in the expansion process. Furthermore, microarray data clearly show that expression levels of AtEXLA2 are differentially regulated during the process of dark-grown hypocotyl elongation. In the early phase of hypocotyl elongation at 48 h post-induction (hpi) AtEXLA2 is 1·7-fold up-regulated compared with 45 hpi. Later during hypocotyl elongation, AtEXLA2 levels keep rising, with a 3·0- and 4·2-fold increase at 52 and 55 hpi, respectively, compared with the onset of elongation at 45 hpi (Pelletier et al., 2010). These transcriptomic data also point towards a role for AtEXLA2 during hypocotyl elongation as its expression rises at the moment when rapid elongation of etiolated hypocotyl cells is seen. As suggested by these data, AtEXLA2 over-expression indeed caused a small but significant growth-promoting effect in the root and etiolated hypocotyl, indicating that increased AtEXLA2 stimulates elongation in the early stages of root and etiolated hypocotyl growth since differences are already apparent in 3-day-old organs.
Strong AtEXLA2 expression was also localized in the central cylinder of the root, leaves and the filament, suggesting that AtEXLA2 could play a role in secondary cell wall deposition in xylem cells. Over-expression of AtEXLA2 would increase the CBDs present in cells, which could interfere somehow with the massive cellulose deposition that is typical for secondary wall formation. This mechanism could explain the decrease in cell wall strength as a result of AtEXLA2 over-expression (Table 1).
It is generally believed that expansins facilitate in vitro cell wall creep and that they therefore are key regulators of cell expansion. In this light, it is important to mention that numerous papers have indeed pointed to the correlation between expansin expression and cellular elongation (e.g. Cho and Kende, 1997; Cho and Cosgrove, 2000; Lee et al., 2003), but several other studies have also stressed that this correlation is far from absolute (Caderas et al., 2000; Rochange et al., 2001; Choi et al., 2003; Sloan et al., 2009). Using transgenic arabidopsis plants with inducible over-expression of cucumber expansin, Goh et al. (2014) have recently proposed that the effects of expansins on growth largely depend on the developmental pattern, organ and the level of changes in expansin expression, and that the effects can vary from growth promotion even to an inhibition. These findings indicate that the levels of expansin in the cell wall are crucial for the resulting phenotypic effect. Under natural conditions, expansins are present in the cell wall at very low concentrations of around one protein per 5000 parts of cell wall (data from cucumber hypocotyls) (McQueen-Mason and Cosgrove, 1995; Cosgrove, 2000b). It is thus not surprising that abnormally high concentrations of these proteins can lead to unexpected phenotypes. From our results, it is clear that expansin-like proteins do not induce drastic changes in cell wall mechanics, as expansins do, so they could have a different mechanism of action.
Besides possible effects of (unnatural) changes in protein abundance in the cell wall, the effects of the over-expression combined with the expression patterns point to a pivotal role for the CBD, especially in the absence of a conventional expansin active site. It is noteworthy that the structure of the EXLA CBD differs slightly from that of α- and β-expansins by harbouring an additional tryptophan and two cysteines. In this light, it is important to mention that over-expression of a CBD-truncated form of swollenin, an expansin-like protein from Trichoderma reesei, did not increase cucumber root colonization, while over-expression of the native form did (Brotman et al., 2008). Also the bacterial CBD of Clostridium cellulovorans has been shown to affect elongation of roots, pollen tubes and root hairs in vitro (Shpigel et al., 1998). Bacterial CBDs have been extensively studied with the aim of changing plant fibre properties and cell wall digestibility (Levy et al., 2002). The effects of over-expression of several CBDs on plant development and cell wall structure has been investigated, and these studies have indicated that the relationship between increased CBD levels and phenotypes is quite ambiguous. For example, over-expression of a tandem fusion protein of CBM29 (carbohydrate-binding module 29) resulted in a reduced stem elongation phenotype (Obembe et al., 2007), while over-expression of a bacterial CBM3 stimulated growth in potato plants (Safra-Dassa et al., 2006). Over-expression of the CBD of the fungus Aspergillus japonicus in arabidopsis also resulted in smaller and thinner plants (van der Valk et al., 2003). Further, it has been shown that some bacterial CBDs are capable of modifying plant cell wall structure in planta (Shpigel et al., 1998; Levy et al., 2002). CBDs are believed to exert their effect on cell walls by influencing normal cell wall polymer organization. In vivo, CBDs are thought to bind to nascent cellulose chains, inhibiting their aggregation into crystalline cellulose microfibrils by shielding the potential hydrogen-bonding sites of the cellulose. Moreover, Shpigel et al. (1998) have reported that increased CBD levels compete for cellulose-binding sites with xyloglucan and result in a lower degree of xyloglucan–cellulose association. Alternatively, CBDs could also bind to cellulose after its synthesis and crystallization, leading to non-hydrolytic fibre disruption (Levy et al., 2002). Also, several other cellulose-binding compounds influence cellulose crystallization, such as fluorescent brighteners (e.g. calco-fluor white) or carboxymethyl cellulose, resulting in cell wall changes in bacteria. The CBD from Clostridium cellulovorans, for example, led to an increased cell wall biosynthesis in Acetobacter xylinum (Shpigel et al., 1998). These documented effects on cellulose and/or cellulose/xyloglucan could explain the observed decrease in the wall strength (Table 1) as a result of AtEXLA2 over-expression.
In bacterial cellulases, CBDs are responsible for the concentration of the catalytic domain at the substrate surface. For some of these cellulases with multiple CBDs, it has been demonstrated that the individual CBDs display different affinities towards specific cell wall polymers. This indicates that the substrate specificity of CBDs largely determines its effect on the cell wall. As most CBDs (or CBMs in general) have been determined based on sequence similarity, their substrate specificities have not yet been proven. Therefore, it is of major importance to determine empirically the binding preferences of the many different CBDs.
Taken together, all these data point to a potential influence of CBDs on cell walls. Cellulose has been proposed as the substrate that is recognized by the CBD of expansins (Cosgrove, 2000b, c). However, the CBD of AtEXLA2 displays some differences when compared with the well-studied α- and β-expansins, potentially reflecting a different substrate specificity. Only when these unknown specificities are elucidated can their exact function and effects in the cell wall be properly interpreted. From the present data, an increase in typical expansin activity by over-expressing AtEXLA2 cannot explain the observed effect on growth. Based on our results, we hypothesize that AtEXLA2 functions in cell wall remodelling, although its exact mechanism of action remains to be determined. It would be very interesting to test whether members of the expansin-like A or B families have any effects on cell wall extension in in vitro cell wall extension assays under a range of protein concentrations and to examine the specificity of their CBDs in binding to cellulose and other cell wall polymers.
SUPPLEMENTARY DATA
Supplementary data are available online at www.aob.oxfordjournals.org and consist of the following. Figure S1: expression patterns of AtEXLA2 in different above-ground organs of arabidopsis revealed by the eFP browser (Winter et al., 2007). Figure S2: expression patterns of AtEXLA2 in the arabidopsis root revealed by the eFP browser (Winter et al., 2007)
ACKNOWLEDGEMENTS
The authors acknowledge Professor Blust (Biology Department University of Antwerp) for the use of the qPCR machinery, and Professor Schryvers for the use of the transmission electron microscope. This work was supported by a PhD grant of the Institute for the Promotion of Innovation through Science and Technology in Flanders (IWT, Vlaanderen); the University of Antwerp; the Research Foundation-Flanders (FWO); the Russian Foundation for Basic Research [14-04-01624a]; and Saint-Petersburg State University [1.23.74.2014 and 1.38.233.2014].
LITERATURE CITED
- Abramoff MD, Magelhaes PJ, Ram SJ. 2004. Image processing with ImageJ. Biophotonics International 11: 36–42. [Google Scholar]
- Baccelli I, Luti S, Bernardi R, Scala A, Pazzagli L. 2014. Cerato-platanin shows expansin-like activity on cellulosic materials. Applied Microbiology and Biotechnology 98: 175–84. [DOI] [PubMed] [Google Scholar]
- Belfield EJ, Ruperti B, Roberts JA, McQueen-Mason S. 2005. Changes in expansin activity and gene expression during ethylene-promoted leaflet abscission in Sambucus nigra. Journal of Experimental Botany 56: 817–823. [DOI] [PubMed] [Google Scholar]
- Blancaflor EB. 2002. The cytoskeleton and gravitropism in higher plants. Journal of Plant Growth Regulation 21: 120–36. [DOI] [PubMed] [Google Scholar]
- Brotman Y, Briff E, Viterbo A, Chet I. 2008. Role of swollenin an expansin-like protein from Trichoderma in plant root colonization. Plant Physiology 147: 779–789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brummell DA, Howie WJ, Ma C, Dunsmuir P. 2002. Postharvest fruit quality of transgenic tomatoes suppressed in expression of a ripening-related expansin. Postharvest Biology and Technology 25: 209–220. [Google Scholar]
- Caderas D, Muster M, Vogler H, et al. 2000. Limited correlation between expansin gene expression and elongation growth rate. Plant Physiology 123: 1399–1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpita NC, Gibeaut DM. 1993. Structural models of primary cell walls in flowering plants: consistency of molecular structure with the physical properties of the walls during growth. The Plant Journal 3: 1–30. [DOI] [PubMed] [Google Scholar]
- Caturla M, Chaparro C, Schroeyers K, Holsters M. 2002. Suppression subtractive hybridization to enrich low-abundance and submergence-enhanced transcripts of adventitious root primordia of Sesbania rostrata. Plant Science 162: 915–921. [Google Scholar]
- Civello PM, Powell ALT, Sabehat A, Bennett AB. 1999. An expansin gene expressed in ripening strawberry fruit. Plant Physiology 121: 1273–1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho HT, Cosgrove DJ. 2000. Altered expression of expansin modulates leaf growth and pedicel abscission in Arabidopsis thaliana. Proceedings of the National Academy of Sciences, USA 97: 9783–9788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho HT, Cosgrove DJ. 2002. Regulation of root hair initiation and expansin gene expression in Arabidopsis. The Plant Cell 14: 3237–3253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho HT, Kende H. 1997. Expansins and internodal growth of deepwater rice. Plant Physiology 113: 1145–1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi DS, Lee Y, Cho HT, Kende H. 2003. Regulation of expansin gene expression affects growth and development in transgenic rice plants. The Plant Cell 15: 1386–1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi D, Cho HT, Lee Y. 2006. Expansins: expanding importance in plant growth and development. Physiologia Plantarum 126: 511–518. [Google Scholar]
- Choi D, Kim JH, Lee Y. 2008. Expansins in plant development. Advances in Botanical Research 47: 47–97. [Google Scholar]
- Cleland R. 1967. Extensibility of isolated cell walls: measurement and changes during cell elongation. Planta 74: 197–209. [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. 1998. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. The Plant Journal 16: 735–743. [DOI] [PubMed] [Google Scholar]
- Cosgrove DJ. 2000a. Expansive growth of plant cell walls. Plant Physiology and Biochemistry 38: 109–124. [DOI] [PubMed] [Google Scholar]
- Cosgrove DJ. 2000b. Loosening of plant cell walls by expansins. Nature 407: 321–326. [DOI] [PubMed] [Google Scholar]
- Cosgrove DJ. 2000c. New genes and new biological roles for expansins. Current Opinion in Plant Biology 3: 73–78. [DOI] [PubMed] [Google Scholar]
- Cosgrove DJ. 2005. Growth of the plant cell wall. Nature Reviews Molecular and Cellular Biology 6: 850–861. [DOI] [PubMed] [Google Scholar]
- Cosgrove DJ, Bedinger P, Durachko DM. 1997. Group I allergens of grass pollen as cell wall-loosening agents. Proceedings of the National Academy of Sciences, USA 94: 6559–6564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dal Santo S, Vannozzi A, Tornielli GB, et al. 2013. Genome-wide analysis of the expansin gene superfamily reveals grapevine-specific structural and functional characteristics. PLoS One 8: e62206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darley CP, Li Y, Schaap P, McQueen-Mason SJ. 2003. Expression of a family of expansin-like proteins during the development of Dicyostelium discoideum. FEBS Letters 546: 416–418. [DOI] [PubMed] [Google Scholar]
- Edwards K, Johnstone C, Thompson C. 1991. A simple and rapid method for the preparation of plant genomic DNA for PCR analysis. Nucleic Acids Research 19: 1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estelle MA, Somerville C. 1987. Auxin-resistant mutants of Arabidopsis thaliana with an altered morphology. Molecular Genomics and Genetics 206: 200–206. [Google Scholar]
- Fonseca S, Monteiro L, Barreiro MG, Pais MS. 2005. Expression of genes encoding cell wall modifying enzymes is induced by cold storage and reflects changes in pear fruit texture. Journal of Experimental Botany 56: 2029–2036. [DOI] [PubMed] [Google Scholar]
- Fry SC, Smith RC, Renwick KF, Martin DJ, Hodge SK, Matthews KJ. 1992. Xyloglucan endotransglycosylase, a new wall-loosening enzyme activity from plants. Biochemical Journal 282: 821–828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Q, Zhao MR, Li F, Guo QF, Xing SC, Wang W. 2008. Expansins and coleoptile elongation in wheat. Protoplasma 233: 73–81. [DOI] [PubMed] [Google Scholar]
- Goh H-H, Sloan J, Malinowski R, Fleming A. 2014. Variable expansin expression in Arabidopsis leads to different growth responses. Journal of Plant Physiology 171: 329–339. [DOI] [PubMed] [Google Scholar]
- Higo K, Ugawa Y, Iwamoto M, Korenaga T. 1999. Plant cis-acting regulatory DNA elements (PLACE) database. Nucleic Acids Research 27: 297–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou G, Kramer VL, Wang YS, et al. 2004. The promotion of gravitropism in Arabidopsis roots upon actin disruption is coupled with the extended alkalinization of the columella cytoplasm and a persistent lateral auxin gradient. The Plant Journal 39: 113–125. [DOI] [PubMed] [Google Scholar]
- Ishikawa H, Evans ML. 1997. Novel software for analysis of root gravitropism: comparative response patterns of Arabidopsis wild-type and axr1 seedlings. Plant, Cell and Environment 20: 919–928. [DOI] [PubMed] [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW. 1987. GUS fusions: β-glucuronidase as a sensitive and versatile gene fusion marker in higher-plants. EMBO Journal 6: 3901–3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones L, McQueen-Mason S. 2004. A role for expansins in dehydration and rehydration of the resurrection plant Craterostigma plantagineum. FEBS Letters 559: 61–65. [DOI] [PubMed] [Google Scholar]
- Karimi M, Inzé D, Depicker A. 2002. GATEWAY™ vectors for Agrobacterium-mediated plant transformation. Trends in Plant Science 7: 193–195. [DOI] [PubMed] [Google Scholar]
- Karimi M, Depicker A, Hilson P. 2007. Recombinational cloning with plant gateway vectors. Plant Physiology 145: 1144–1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kende H, Kent J, Bradford KJ, et al. 2004. Nomenclature for members of the expansin superfamily of genes and proteins. Plant Molecular Biology 55: 311–314. [DOI] [PubMed] [Google Scholar]
- Kerff F, Amoroso A, Herman RI, et al. 2008. Crystal structure and activity of Bacillus subtilis YoaJ (EXLX1), a bacterial expansin that promotes root colonization. Proceedings of the National Academy of Sciences, USA 105: 16876–16881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koncz C, Schell J. 1986. The promoter of TL-DNA gene 5 controls the tissue-specific expression of chimeric genes carried by a novel type of Agrobacterium binary vector. Molecular Genomics and Genetics 204: 383–396. [Google Scholar]
- Kudla U, Qin L, Milac A, et al. 2005. Origin distribution and 3D-modeling of Gr-EXPB1 an expansin from the potato cyst nematode Globodera rostochiensis. FEBS Letters 579: 2451–2457. [DOI] [PubMed] [Google Scholar]
- Labrador E, Nevins DJ. 1989. An exo-/β-d-glucan derived from Zea coleoptile walls with a capacity to elicit cell elongation. Physiologia Plantarum 77: 479–486. [Google Scholar]
- Lee DK, Ahn JH, Song SK, Choi YD, Lee JS. 2003. Expression of an expansin gene is correlated with root elongation in soybean. Plant Physiology 131: 985–997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HJ, Lee S, Ko HJ, Kim KH, Choi IG. 2010. An expansin-like protein from Hahella chejuensis binds cellulose and enhances cellulase activity. Molecules and Cells 29: 379–85. [DOI] [PubMed] [Google Scholar]
- Lescot M, Dehais P, Thijs G, et al. 2002. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Research 30: 325–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy I, Shani Z, Shoseyov O. 2002. Modification of polysaccharides and plant cell wall by endo-1,4-beta-glucanase and cellulose-binding domains. Biomolecular Engineering 19: 17–30. [DOI] [PubMed] [Google Scholar]
- Li Y, Darley CP, Ongaro V, et al. 2002. Plant expansins are a complex multigene family with an ancient evolutionary origin. Plant Physiology 128: 854–864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Jones L, McQueen-Mason S. 2003. Expansins and cell growth. Current Opinion in Plant Biology 6: 603–610. [DOI] [PubMed] [Google Scholar]
- Maris A, Kaewthai N, Eklöf JM, et al. 2011. Differences in enzymic properties of five recombinant xyloglucan endotransglucosylase/hydrolase (XTH) proteins of Arabidopsis thaliana. Journal of Experimental Botany 62: 261–271. [DOI] [PubMed] [Google Scholar]
- McQueen-Mason S, Cosgrove DJ. 1994. Disruption of hydrogen-bonding between plant-cell wall polymers by proteins that induce wall extension. Proceedings of the National Academy of Sciences, USA 91: 6574–6578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McQueen-Mason SJ, Cosgrove DJ. 1995. Expansin mode of action on cell walls – analysis of wall hydrolysis stress-relaxation and binding. Plant Physiology 107: 87–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McQueen-Mason S, Durachko DM, Cosgrove DJ. 1992. Two endogenous proteins that induce cell-wall extension in plants. The Plant Cell 4: 1425–1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miedes E, Suslov D, Vandenbussche F, et al. 2013. Xyloglucan endotransglucosylase/hydrolase (XTH) overexpression affects growth and cell wall mechanics in etiolated Arabidopsis hypocotyls. Journal of Experimental Botany 64: 2481–2497. [DOI] [PubMed] [Google Scholar]
- Miller ND, Parks BM, Spalding EP. 2007. Computer-vision analysis of seedling responses to light and gravity. The Plant Journal 52: 374–81. [DOI] [PubMed] [Google Scholar]
- Nakagawa T, Kurose T, Hino T, et al. 2007. Development of series of gateway binary vectors, pGWBs, for realizing efficient construction of fusion genes for plant transformation. Journal of Bioscience and Bioengineering 104: 34–41. [DOI] [PubMed] [Google Scholar]
- Nardi C, Escudero C, Villarreal N, Martinez G, Civello PM. 2013. The carbohydrate-binding module of Fragaria×ananassa expansin 2 (CBM-FaExp2) binds to cell wall polysaccharides and decreases cell wall enzyme activities ‘in vitro’. Journal of Plant Research 126: 151–159. [DOI] [PubMed] [Google Scholar]
- Nieuwland J, Feron R, Huisman BAH, et al. 2005. Lipid transfer proteins enhance cell wall extension in tobacco. The Plant Cell 17: 2009–2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishitani K, Vissenberg K. 2007. Roles of the XTH protein family in the expanding cell. In: Verbelen J-P, Vissenberg K, eds. The expanding cell. Plant Cell Monographs, vol. 5 Berlin: Springer, 89–116. [Google Scholar]
- Obembe OO, Jacobsen E, Timmers J, et al. 2007. Promiscuous, non-catalytic, tandem carbohydrate-binding modules modulate the cell-wall structure and development of transgenic tobacco (Nicotiana tabacum) plants. Journal of Plant Research 120: 605–617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto-Nakazato A, Takahashi K, Katoh-Semba R, Katou K. 2001. Distribution of yieldin a regulatory protein of the cell wall yield threshold in etiolated cowpea seedlings. Plant and Cell Physiology 42: 952–958. [DOI] [PubMed] [Google Scholar]
- Orford SJ, Timmis JN. 1998. Specific expression of an expansin gene during elongation of cotton fibres. Biochima et Biophysica Acta 1398: 342–346. [DOI] [PubMed] [Google Scholar]
- Passardi F, Tognolli M, De Meyer M, Penel C, Dunand C. 2006. Two cell wall associated peroxidases from Arabidopsis influence root elongation. Planta 223: 965–974. [DOI] [PubMed] [Google Scholar]
- Pelletier S, Van Orden J, Wolf S, et al. 2010. A role for pectin de-methylesterification in a developmentally regulated growth acceleration in dark-grown Arabidopsis hypocotyls. New Phytologist 188: 726–739. [DOI] [PubMed] [Google Scholar]
- Pfaffl MW. 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Research 29: e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin L, Kudla U, Roze EHA, et al. 2004. Plant degradation: a nematode expansin acting on plants. Nature 427: 30. [DOI] [PubMed] [Google Scholar]
- Rayle DL, Cleland RE. 1992. The acid growth theory of auxin-induced cell elongation is alive and well. Plant Physiology 99: 1271–1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rochange SF, Wenzel CL, McQueen-Mason SJ. 2001. Impaired growth in transgenic plants over-expressing an expansin isoform. Plant Molecular Biology 46: 581–589. [DOI] [PubMed] [Google Scholar]
- Roland DR, Mosiniak M, Vian B. 1982. Cell wall texture along the growth gradient of the mung bean hypocotyl: ordered assembly and dissipative processes. Journal of Cell Science 56: 303–318. [Google Scholar]
- Rombauts S, Dehais P, Van Montagu M, Rouze P. 1999. PlantCARE, a plant cis-acting regulatory element database. Nucleic Acids Research 27: 295–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Safra-Dassa L, Shani Z, Danin A, Roiz L, Shoseyov O, Wolf S. 2006. Growth modulation of transgenic potato plants by heterologous expression of bacterial carbohydrate-binding module. Molecular Breeding 17: 355–364. [Google Scholar]
- Sakurai T, Satou M, Akiyama K, et al. 2005. RARGE: a large-scale database of RIKEN Arabidopsis resources ranging from transcriptome to phenome. Nucleic Acids Research 33: D647–D650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampedro J, Cosgrove DJ. 2005. Protein family review: the expansin superfamily. Genome Biology 6: 242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheres B, Wolkenfelt H, Willemsen V, et al. 1994. Embryonic origin of the Arabidopsis primary root and root meristem initials. Development 120: 2475–2487. [Google Scholar]
- Scholl RL, May ST, Ware DH. 2000. Seed and molecular resources for Arabidopsis. Plant Physiology 124: 1477–1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seki M, Carninci P, Nishiyama Y, Hayashizaki Y, Shinozaki K. 1998. High-efficiency cloning of Arabidopsis full-length cDNA by biotinylated CAP trapper. The Plant Journal 15: 707–720. [DOI] [PubMed] [Google Scholar]
- Seki M, Narusaka M, Kamiya A, et al. 2002. Functional annotation of a full-length Arabidopsis cDNA collection. Science 296: 141–145. [DOI] [PubMed] [Google Scholar]
- Shcherban TY, Shi J, Durachko DM, et al. 1995. Molecular cloning and sequence analysis of expansins – a highly conserved, multigene family of proteins that mediate cell wall extension in plants. Proceedings of the National Academy of Sciences, USA 92: 9245–9249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shpigel E, Roiz L, Goren R, Shoseyov O. 1998. Bacterial cellulose-binding domain modulates in vitro elongation of different plant cells. Plant Physiology 117: 1185–1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloan J, Backhaus A, Malinowski R, McQueen-Mason S, Fleming AJ. 2009. Phased control of expansin activity during leaf development identifies a sensitivity window for expansin-mediated induction of leaf growth. Plant Physiology 151: 1844–1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suslov D, Verbelen J-P. 2006. Cellulose orientation determines mechanical anisotropy in onion epidermis cell walls. Journal of Experimental Botany 57: 2183–2192. [DOI] [PubMed] [Google Scholar]
- Thompson JE, Fry SC. 2001. Restructuring of wall-bound xyloglucan by transglycosylation in living plant cells. The Plant Journal 26: 23–34. [DOI] [PubMed] [Google Scholar]
- van der Valk H, Quentin M, van Dam J, de Jong E. 2003. Cellulose-binding domains: tools for innovation in cellulose fibre production and modification. ACS Symposium Series 855: 132–155. [Google Scholar]
- Vandenbussche F, Suslov D, De Grauwe L, Leroux O, Vissenberg K, Van Der Straeten D. 2011. The role of brassinosteroids in shoot gravitropism. Plant Physiology 156: 1331–1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Sandt VST, Suslov D, Verbelen J-P, Vissenberg K. 2007. Xyloglucan endotransglucosylase activity loosens a plant cell wall. Annals of Botany 100: 1467–1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Tang R, Tao J, et al. 2011. Quantitative investigation of non-hydrolytic disruptive activity on crystalline cellulose and application to recombinant swollenin. Applied Microbiology and Biotechnology 91: 1353–1363. [DOI] [PubMed] [Google Scholar]
- Winter D, Vinegar B, Nahal H, Ammar R, Wilson GV, Provart NJ. 2007. An ‘Electronic Fluorescent Pictograph’ browser for exploring and analyzing large-scale biological data sets. PLoS One 2: e718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu BZ, Janson JC, Sellos D. 2001. Cloning and sequencing of a molluscan endo-beta-1 4-glucanase gene from the blue mussel Mytilus edulis. European Journal of Biochemistry 268: 3718–3727. [DOI] [PubMed] [Google Scholar]
- Young LS, Harrison BR, Narayana Murthy UM, Moffatt BA, Gilroy S, Masson PH. 2006. Adenosine kinase modulates root gravitropism and cap morphogenesis in Arabidopsis. Plant Physiology 142: 564–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann P, Hirsch-Hoffmann M, Hennig L, Gruissem W. 2004. GENEVESTIGATOR. Arabidopsis microarray database and analysis toolbox. Plant Physiology 136: 2621–2632. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.