Abstract
Background and Aims Quercus suber (cork oak) is a dominant tree of the Fagaceae in forests of the south-west Iberian Peninsula. It is monoecious with a long progamic phase that provides a comprehensive system for comparative studies in development and sexual reproduction. In this study the distribution of arabinogalactan protein (AGPs) and pectin epitopes in anthers of Q. suber was assessed to map these hydroxyproline-rich glycoproteins and the galacturonate-rich acidic polysaccharides during pollen development.
Methods Immunolocalization in male flowers was performed with a set of monoclonal antibodies directed against the carbohydrate moiety that recognizes AGPs and pectins. To identify AGP genes involved in cork oak male flower development, a search was conducted for annotated AGP genes in the available transcriptome data of the Cork Oak EST Consortium database (www.corkoakdb.org).
Key Results Ubiquitous labelling in all cell types was obtained with anti-homogalacturan antibodies for methyl-esterified pectins. In contrast, the antibody that labelled non-methyl-esterified homogalacturans had a preferential presence in microsporocyte cells walls at the beginning of pollen development. Intense labelling was obtained with anti-AGP antibodies both in the tapetum and in the intine wall near the pollen apertures and later in the generative cell wall and vegetative cell. Evaluation of the putative AGPs highly expressed in the male gametophyte was achieved by quantitative RT-PCR analysis in male and female cork oak flowers.
Conclusions Four putative AGP genes were identified that are preferentially expressed in the male flower compared with the female flower. The putative Arabidopsis thaliana orthologues of these genes are associated with preferential expression in pollen, suggesting that the AGPs probably play a significant role in cork oak reproduction.
Keywords: Quercus suber, cork oak, pollen development, arabinogalactan proteins, AGP, pectins, microsporocyte cells walls, male gametophyte, Arabidopsis thaliana
INTRODUCTION
In higher plants, pollen development is of major biological importance for the reproductive success of most species. Pollination success is also commercially important as it determines crop fertility and consequent productivity. Although the mechanism of pollen development has been described for several plant species, little is known about the complex regulatory networks that control the transition from a sporophytic to a gametophytic phase of development. Several recent studies have revealed many different genes that play roles in pollen development in different species, especially in the model plant Arabidopsis thaliana (Twell, 2011). Arabinogalactan proteins (AGPs) have been implicated in different processes of plant growth and development such as somatic embryogenesis, root development, hormone responses and programmed cell death (Seifert and Roberts, 2007). However, AGPs have been closely associated with reproductive function not only because they are mainly expressed in sexual reproductive tissues but also because they may play an important role in anther and pollen development (Ma and Sundaresan, 2010; Costa et al., 2013a). AGPs are a class of hydroxyprolin-rich glycoproteins, ubiquitous in the plant kingdom, that are expressed in almost all types of cells. AGPs are either present at the cell surface anchored to the plasma membrane, as part of the cell wall, or as extracellular secretions. AGPs are probably the most highly modified proteins in nature undergoing several post-translational modifications. More than 90 % of the molecule is made up of sugars, hampering study of this protein type. AGPs are classified into four classes based on the protein backbone; classical AGPs have an N-terminal signal peptide, a central domain rich in proline/hydroxyproline and a C-terminal domain containing a glycosylphosphatidyl inositol (GPI) anchor signal sequence. Other AGP types are lysine-rich, with a lysine-rich domain, which may play crucial roles in cell communication and adhesion, AG peptides with short protein backbones and fasciclin-like AGPs with one or more fasciclin domains (Schultz et al., 2002; Johnson et al., 2003).
AGPs comprise a class of proteins with several homologues, and in arabidopsis at least 85 AGP genes are known (Showalter et al., 2010). AGPs have also been shown to be differentially expressed during gametophyte development, making them important molecular markers for reproductive development (Coimbra et al., 2007, 2009). Due to their molecular constituents and a prevailing localization in female and male flower tissues, such as the stigma exudates and transmission tract, and in the pollen grain and pollen tube, they are believed to provide guidance clues and attraction signals for pollen tube guidance into the embryo sac (Costa et al., 2013b). As in arabidopsis, rice and apple AGP-encoding genes also control pollen function (Coimbra et al., 2007, 2009; Levitin et al., 2008; Ma and Zhao, 2010). The evergreen cork oak (Quercus suber) was chosen to further study the involvement of AGPs in male flower reproductive development. Quercus suber is a Fagaceae tree species which dominates the forests of the southern Iberian Peninsula. It is one of the most important forest species in Portugal due to its ecological and socio-economic significance. Not only is there a keen interest in this species because of cork production but also there is a growing interest in the production of acorns destined either for nursery production or for animal feed stocks. In the last few years, considerable effort has been made to study the biological mechanisms associated with this species (Pereira-Leal et al., 2014), and therefore knowledge of the molecular mechanisms that control flower development is crucial to fully understand the reproductive success of this species.
Like Arabidopsis thaliana, Q. suber is a core eudicot species, suggesting late divergence, although the reproductive habit of the two species is very different, indicating the rapid adaptation of the angiosperms to different environments. The question that remains to be answered is if the species adaptation is based on the establishment of new traits or the adaptation of old ones. Cork oak is a monoecious species in which female flowering buds appear in spring, whereas male flowers occur in early spring and sometimes in autumn. Female flowers are partially enclosed by a dome-shaped bud of imbricate scales and male staminate flowers occur in catkins at the base of current season branches. At anthesis, female flowers are not completely developed. Ten to 12 d after pollination, pollen tube growth is arrested, and its growth is only resumed around 3 months after pollination (Boavida et al., 1999). Such characteristics make Q. suber an interesting system for comparative studies of development and sexual reproduction in a non-model forest plant. As mentioned, development of the female gametophyte occurs only after a delayed process of pollination, and the interactions between the pollen tube and the pistil tissues may involve different players (Boavida et al., 2001). Therefore, it is possible that AGPs could be involved in this process. The lack of a complete sequenced genome has restricted genetic studies on this plant, but recently a National Cork Oak Consortium has produced and sequenced representative expressed sequence tags (ESTs) from cork oak (Pereira-Leal et al., 2014). In the present study, the data generated by the National Cork Oak Consortium were used to determine the putative involvement of AGP in male reproductive organ development. Because AGPs are known to be involved in pollen development and in pollen–pistil interactions, in the present work we also look at the tissue and stage development specificity of AGP epitopes, recognized by monoclonal antibodies (Mabs). This work will not only shed light on the evolutive interspecific relationships between different AGPs but also allow a comparative analysis between species with different reproductive strategies. Any knowledge concerning the reproductive biology of Q. suber is essential to understand the molecular mechanisms of seed production and identify any constraints affecting the reproductive success of this species, as oaks have large variations in acorn production from year to year among individual trees (Farmer, 1981).
MATERIAL AND METHODS
Plant material and light microscopy
Individual flowers from male inflorescences of Quercus suber, collected from randomly selected trees of two natural populations in the Porto area, were fixed in 2 % (w/v) paraformaldehyde and 2·5 % (w/v) glutaraldehyde in phosphate buffer [0·025 m, pH 7, 0·001 % (v/v) Tween 80], placed under vacuum for 1 h and then at 4 °C overnight. After dehydration in a graded ethanol series, the material was embedded in LR White. Thick sections (0·5 µm) were obtained with a Leica Reichert Supernova microtome, placed on glass slides and stained with a solution of 1 % (w/v) toluidine blue (Sigma-Aldrich, St Louis, MO, USA) or 1% (w/v) Lugol solution (Sigma-Aldrich 62650) for starch staining. Histology mounting medium Fluoroshield™ with DAPI (F6057 Sigma) was used to detect the nucleus in the pollen grains. Some slides were left unstained for immunolocalization with Mabs against AGPs and pectin epitopes. Sections for brightfield microscopy were mounted with Eukitt quick-hardener (Fluka).
Immunolocalization and antibodies
A selection of Mabs directed against specific AGPs and pectin were provided by Professor Paul Knox from the Centre for Plant Sciences, University of Leeds, UK. The Mabs against AGPs used were JIM8 (Pennell et al., 1991), JIM13 (Knox et al., 1991), MAC207 (Pennell et al., 1989) and LM6 (which also recognizes a type I rhamnogalacturonan) (Willats et al., 1998) and for pectins LM5 (Jones et al., 1997), JIM5 (Willats et al, 2000) and JIM7 (Knox et al, 1990). Fluorescein isothiocyanate (FITC)-conjugated anti-rat IgG (Sigma-Aldrich F-1763) was used as secondary antibody.
In the slides prepared for immunolocalization, the sections were circled with a PAP pen (Sigma-Aldrich Z672548), and treated as follows: 5 min in phosphate-buffered saline (PBS), pH 7·4, containing 5 % (w/v) non-fat dried milk (blocking solution), followed by incubation with primary antibody (diluted 1 : 5 in blocking solution), for 2 h at room temperature followed by overnight at 4 °C. After washing with PBS, the sections were incubated with secondary antibody (diluted 1 : 100 in blocking solution) for 4 h in the dark, and then finally washed with PBS followed by distilled water. Slides were further stained with 0·01 % (w/v) calcofluor white (Fluorescent Brightener 28; Sigma-Aldrich F3543) and mounted with Vectashield mounting medium (Vector Laboratories, Burlingame, CA, USA; ref: H-1000). Control experiments were performed omitting the incubation with the primary antibody (incubation with blocking solution only), and resulted in no staining with secondary antibody.
Brightfield and fluorescence observations were performed on a Leica DMLB epifluorescence microscope (objectives were Leica N-Plan, and the filters used were 365/445 nm for calcofluor and DAPI; 470/525 nm for fluorescein stain). Images were captured with a ProgRes® MF cool (Jenoptik, Jena, Germany) in automatic exposure mode, and processed with ProgRes® CapturePro 2.8.8 software.
Identification of putative AGP-like genes in Q. suber
Several A. thaliana AGP protein sequences (Table 1) were used as query in a BLASTp analysis using the BLAST tool in the Cork Oak EST Consortium database (www.coarkoakdb.org.). The Quercus AGP-retrieved sequences were aligned with their putative homologues in A. thaliana using BLASTP2.2.29 (Stephen et al., 2005) and each was labelled according to the maximum similarity score obtained with the putative arabidopsis homologues (Table 1). Signal peptides were identified using the SignalP 4.1 server (http://www.cbs.dtu.dk/services/SignalP/) (Petersen et al., 2011), and GPI lipid anchor sequences were identified using the big-PI Plant Predictor (http://mendel.imp.ac.at/gpi/plant_server.html) (Eisenhaber et al., 2003).
Table 1.
Arabidopsis thaliana AGP TAIR and Q. suber Cork Oak database accession numbers
| Species/gene | Accession no. |
|---|---|
| Quercus suber | |
| QsAGP20L1 | QS041483.0 |
| QsAGP20L3 | QS014269.0 |
| QsAGP16L1 | QS001499.0 |
| QsAGP16L2.1 | QS076008.0 |
| QsAGP23 | QS132451.0 |
| QsAGP15 | QS124091.0 |
| QsFLA8 | QS111406.0 |
| QsUBQ | QS149226.0 |
| QsPP2AA3 | QS092015.0 |
| Arabidopsis thaliana | |
| AtAGP6 | At5g14380.1 |
| AtAGP11 | At3g01700.1 |
| AtAGP15 | At5g11740.1 |
| AtAGP16 | At2g46330.2 |
| AtAGP20 | At3g61640.1 |
| AtAGP21 | At1g55330.1 |
| AtAGP22 | At5g53250.1 |
| AtAGP23 | At3g57690.1 |
| AtAGP24 | At5g40730.1 |
| AtAGP40 | At3g20865.1 |
| AtAGP1 | At5g64310.1 |
| AtFLA8 | At2g45470.1 |
| AtPP2AA3 | At1g13320.1 |
| AtUBQ1 | At3g52590.1 |
Phylogenetic analysis
AGP full-length amino acid sequences were aligned using Clustal W (Thompson et al., 1994) and the alignments were used as input into MEGA 5 (Tamura et al., 2011) to generate a phylogenetic tree using the neighbour-joining (NJ) method. Bootstraping was performed 1000 times to obtain support values for each branch. Branches corresponding to partitions reproduced in <50 % bootstrap replicates were collapsed. The maximum-parsimony method within MEGA was also used to support the NJ tree. The TAIR (The Arabidopsis Information Resource) accession numbers of arabidopsis peptide sequences used in the reconstruction of the phylogenetic tree are listed in Table 1.
Quantitative RT-PCR analysis
Unisexual flowers from six distinct developmental stages (according to Varela and Valdiviesso, 1996) were harvested between the end of March and the beginning of June and were frozen in liquid nitrogen immediately after collection. The female samples included young female spikes with flowers with closed scales, up to flowers in which all the stigmas had lost receptivity. Similarly, the male samples included compacted young catkins up to flowers with brownish pollen-shedding anthers. Total RNA of each sample was obtained using the CTAB/LiCl extraction method (Chang et al., 1993) with some modifications (Azevedo et al., 2003). Equal amounts of RNA from the different male and female developmental stages were mixed together into two distinct pools (male and female). cDNA was synthesized according to the Invitrogen cDNA synthesis kit SuperScript® III RT (Carlsbad, CA, USA; ref. 18080-051). cDNA was synthesized from 1 μg total RNA according to the manufacturer’s instructions. cDNA was amplified in real-time PCR with 250 nm of each gene-specific primer (listed in Supplementary Data Table S1) using SsoFast™ EvaGreen® Supermix (Bio-Rad, Hercules, CA, USA; ref: 172-5203) on the CFX96 Touch™ Real-Time PCR Detection System (Bio-Rad) and 1 µL of cDNA (1 : 100 dilution). Real-time PCRs were performed in triplicate. After an initial period of 3 min at 95 °C, each of the 40 PCR cycles consisted of a denaturation step of 10 s at 95 °C, and an annealing/extension step of 10 s at the gene-specific temperature. With each PCR, a melting curve was obtained to check for amplification specificity and reaction contaminations, by heating the amplification products from 60 to 95 °C in 5-s intervals. Primer efficiency was analysed with CFX Manager™ Software v3.1 (Bio-Rad), using the Livak calculation method for normalized expression (Livak and Schmittgen, 2001). Gene expression analysis was established based on three technical replicates, and normalized with two Q. suber homologues for the reference genes AtPP2AA3 and AtUBQ1 (Table 1).
RESULTS
Quercus suber microgametogenesis
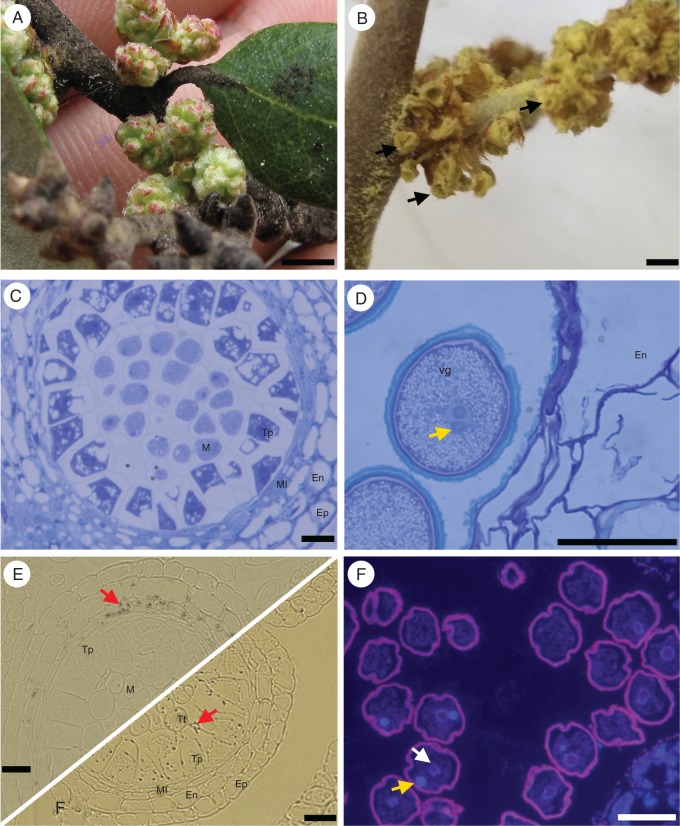
Quercus suber male gametogenesis begins with the bolting of male flowers (Fig. 1A), at which point the immature anthers all have five cell-wall layers: the microsporocytes in the centre are surrounded by a well-developed tapetum, the median layer, the endothecium and the epidermis (Fig. 1C). During this stage of development, starch grains accumulate predominantly along the median layer (Fig. 1E). As the catkins start to elongate, with the anthers still largely covered by bracts, the microsporocyte cells go into meiosis, and the starch grains appear mostly distributed between the recently formed tetrads and the tapetum layer (Fig. 1F). Further in development, anthers emerge from the bracts and lose their green colour (Fig. 1B), and at this point the callose wall that surrounded the tetrads is dissolved and the released microspores undergo the typical asymmetric cell division (Fig. 1D). At anthesis starch was found in small granules in the pollen cytoplasm, and the generative cell, quite small in size, is localized near the vegetative nucleus, and the pollen is shed bicellularly (Fig. 1G).
Fig. 1.
Morphological aspects of Quercus suber microgametogenesis. (A) Young male inflorescence buds in early spring. (B) Mature male flower with dehiscent anthers (arrows). (C) Histological section of a young anther locule, showing the epidermis (Ep), endothecium (En), middle layer (Ml), tapetum (Tp) and microsporocytes (M). (D) Mature pollen grain; the generative cell (arrow) does not undergo mitosis and can usually be seen close to the vegetative cell (Vg) nucleus. (E) Before meiosis, the starch grains (red arrows) accumulate in the middle layer of the anther. (F) After meiosis the starch is detected in the microspores. (G) DAPI staining shows the generative (yellow arrow) and vegetative cell (white arrow) nuclei after asymmetric division of the microspore. Scale bars = 1 cm (A and B); 40 µm (C, D, E, F and G). En, endothecium; Ml, middle layer; Tp, tapetum; M, microsporocyte; Tt, tetrad.
Distribution of homogalacturonans during male gametogenesis
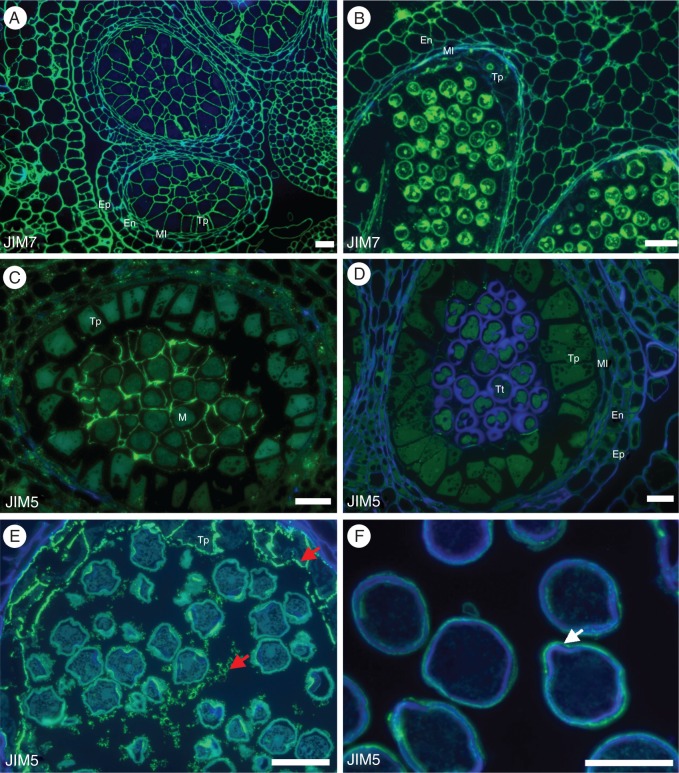
Pectins are one of the main components of the cell-wall matrix. During development pectins that are methyl-esterified can be distinguished from those that are not methyl-esterified by labelling with two different monoclonal antibodies, JIM7 and JIM5, respectively. JIM7 labelling shows a ubiquitous distribution of methyl-esterified homogalacturonan epitopes throughout pollen development, being present in all cell types during all developmental stages independently of cell fate (Fig. 2A, B). JIM5 labelling shows that partially methyl-esterified homogalacturonans have a preferential distribution in the microsporocyte cell walls at the beginning of pollen development (Fig. 2C). These cell walls are very important for development and cell fate, and the labelling is no longer detected as meiosis progresses and tetrads are formed (Fig. 2D). Later in development, this antibody labels the tapetum wall and the vesicles secreted by this tissue (Fig. 2E). Near the end of pollen development, JIM5 epitopes are very faintly present at the pollen grain pores (Fig. 2F).
Fig. 2.
Distribution of homogalacturonans in Quercus suber microgametogenesis. (A) JIM7 labelling of the partially methyl-esterified homogalacturonan at the pre-meiotic stage of pollen development, showing a ubiquitous and constant distribution of epitopes in all pectocellulosic cell walls. (B) JIM7 labelling at the bicellular pollen stage of development shows no distinctive pattern of epitope distribution. (C) Contrasting with JIM7 distribution, the weakly methyl-esterified homogalacturonans labelled by JIM5 showed a more restricted distribution of epitopes at early stages of pollen development, being detected only at the cell walls of the pollen mother cells (M). (D) As meiosis progresses the labelling fades completely. (E) Later only after release of the unicellular microspore, JIM5 labels the vesicles secreted by the tapetum and the tapetum itself. (F) At the mature pollen stage, JIM5 can only be seen in the intin wall next to the pollen pores (arrow). Scale bars = 40 µm. En, endothecium; Ml, middle layer; Tp, tapetum; M, microsporocytes; Tt, tetrad.
Distribution of type I rhamnogalacturonans during Q. suber microgametogenesis
LM6 recognizes α-(1,5)-L-arabinan from a type I rhamnogalacturonan (RG-I) but it also has affinity for AGP arabinans. The epitopes recognized by this antibody were present in all cell walls at the beginning of anther development (Fig. 3A) but showed stronger intensity in tapetum cell walls and microsporocytes as soon as these cells differentiate (Fig. 3B). Labelling intensity decreased and could not be seen at the walls of tetrads or of released microspores. As pollen development continues, this antibody labelled the vesicles secreted by the tapetum and the cytoplasm and intine in microspores (Fig. 3C). After pollen mitosis, the epitopes recognized by LM6 were clearly present at the generative cell wall (Fig. 3D).
Fig. 3.
Type I rhamnogalacturonan distribution during Quercus suber microgametogenesis. (A) At the early stage of anther differentiation LM6 labels all cell walls of the anther. (B) During pre-meiosis of the anther, labelling is strongly associated with the cell wall of the tapetum cells (Tp) and pollen mother cells (M). (C) Later during early bicellular microspore stage of pollen development, LM6 appears to be associated with the tapetum (Tp) membrane and with the vesicles (red arrows) present inside the maturing pollen grain as well and in the vesicles secreted by the tapetum. (D) In the mature pollen grain LM6 labels the intine of the vegetative cell as well as the generative cell (yellow arrow). (E) When LM5, a more specific anti-RG-I Mab is used, labelling is totally absent from the tapetum (Tp) of the pre-meiotic anther. (F) LM5 labelling fades completely inside the locule as the cell wall of the pollen mother cells are degraded during formation of the tetrads. (G) During final maturation of the pollen, LM5 clearly labels the tapetum wall and its secretions to the locule (arrows). (H) In the mature pollen grain, LM5 only faintly labels inside the pollen cytoplasm. Scale bars = 40 µm. En, endothecium; Ml, middle layer; Tp, tapetum; M, microspore.
LM5 recognizes specifically a proportion of RG-I pectins that seemed to have a wide distribution in the pre-meiotic anther with an unexplained exclusion from tapetal cells (Fig. 3E). As meiosis progresses the labelling fades inside the anther locule as tetrads start to differentiate (Fig. 3F). During microspore maturation and pollen mitosis, LM5 signal reappears in the tapetal cells and in its secretions (Fig. 3G), reverting to vestigial levels in all anther tissues at the mature pollen grain stage (Fig. 3H).
AGP distribution during Q. suber male gametogenesis
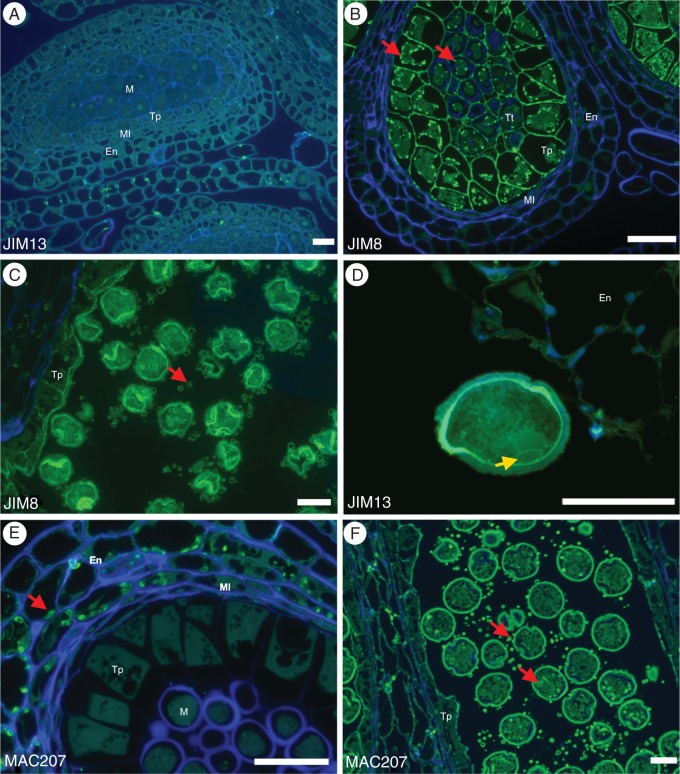
JIM8 and JIM13 were two of the antibodies used against AGP epitopes. These epitopes had an identical distribution pattern, with only a small difference in signal intensity. During early stages of pollen development, the strength of labelling by these antibodies was more intense, especially in the tapetum and in the microsporocytes, cell and walls (Fig. 4A, B). As pollen development continues the labelling remains in tapetal cells and secretions, in the intine wall near the pollen apertures (Fig. 4C) and later specifically in the generative cell wall and vegetative cell intine wall (Fig. 4D).
Fig. 4.
Arabinogalactan protein distribution during Quercus suber microgametogenesis. JIM8 and JIM13 are two anti-arabinogalactan protein Mabs that have the same labelling pattern during Q. suber microgametogenesis. (A) During the initial differentiation stage of pollen mother cells, the AGPs labelled by JIM8 and JIM13 are absent. (B) During meiosis, both these antibodies specifically mark the tapetum and the pollen mother cell walls, but also show strong affinity to the vesicles present in these same cells (red arrows). (C) Later during development, both epitopes are associated with the intine of the microspore and with the membrane of the vesicles secreted by the tapetum (arrow). (D) On the mature pollen grains, JIM8 and JIM13 label both the intine of the vegetative cell and the generative cell wall (arrow). (E) Another AGP-associated monoclonal antibody, MAC207, is associated with the starch granules (red arrows) in Q. suber anther wall layers and has no affinity with the cell walls. (F) In the final stage of pollen maturation, MAC207-associated epitopes can only be seen associated with tapetum secreted vesicles and with the pollen vesicles (arrows). Scale bars = 40 µm. En, endothecium; Ml, middle layer; Tp, tapetum; M, microspore; Tt, tetrad.
MAC207, another anti-AGP antibody, showed a distinct distribution pattern, surrounding starch-rich vesicles at the anther wall layers (Fig. 4E). During pollen maturation, the epitopes recognized by this antibody were also present in tapetal cells and in its secreted vesicles (Fig. 4F).
Putative AGP-like genes in Q. suber related to male gametogenesis
To identify AGP genes involved in cork oak pollen development, we searched for annotated AGP genes in the available transcriptome data of the Cork Oak EST Consortium database (www.corkoakdb.org). Fifty-seven AGP unigenes were found, including four complete genes that encoded putative AG peptide sequences that are AtAGP16 and AtAGP20 homologues. Thus, they were designated QsAGP16-LIKE1 (QsAGP16L1), QsAGP16-LIKE2 (QsAGP16L2), QsAGP20-LIKE1 (QsAGP20L1) and QsAGP20-LIKE3 (QsAGP20L3). Homologues for these putative AG-like peptides were found in Populus trichocarpa, Vitis vinifera, Zea mays and other angiosperms. QsAGP16L1, QsAGP16L2, QsAGP20L1 and QsAGP20L3 peptides are 64, 63, 64 and 78 amino acids long, respectively, and exhibit a PAST domain (proline, alanine, serine and threonine amino acids). They also have an N-terminal signal sequence for targeting to the endoplasmic reticulum where glycosylation occurs, and a hydrophobic C-terminal sequence that is a GPI anchor attachment signal (Supplementary Data Fig. S1). Surprisingly, we found two putative splice variants of the gene QsAGP16L2, namely QsAGP16L2.1 (accession no. QS076008.0) and the QsAGP16L2.2 (accession no. QS007723.0), which encode 63 and 43 residues, respectively. The predicted proteins of these two Q. suber transcripts are 100 % identical throughout their N terminus and core backbone of the AG peptide, with the QsAGPs16L2.2 variant not exhibiting the sequence for GPI-addition anchor.
To obtain additional cork oak AGPs related to pollen development, we used the BLAST program to search the cork oak EST database using as queries AGPs reported as being preferentially or predominantly expressed in arabidopsis pollen (according to Wang et al., 2008). This revealed two putative arabidopsis orthologue genes QsAGP15 and QsAGP23, with 58 and 55 % sequence identity to AtAGP15 and AtAGP23 proteins, respectively. Moreover, both genes encode predicted AG peptide sequences, as both presented the characteristic N- and C-terminal domains and a short PAST-rich peptide, 66 and 72 amino acids long for QsAGP15 and QsAGP23, respectively (Supplementary Data Fig. S1). In particular, QsAGP23 exhibited a core protein of 22 amino acids, consisting of AP, PA and SP (alanine/proline; proline/alanine; serine/proline) repeats and H (histidine) repeats.
A phylogenetic tree was produced to elucidate evolutionary relationships between 11 of the arabidopsis AGPs and the six Q. suber putative AGPs retrieved from the cork oak EST database mentioned above. The phylogenetic tree (Fig. 5A), rooted with FLA8, showed a clade that includes QsAGP23 and most of the arabidopsis AGP pollen-specific genes such as AtAGP11 and AtAGP6.
Fig. 5.

Identification and expression analysis of putative arabidopsis orthologue AGP genes of Quercus suber. (A) NJ tree based on 19 AGP amino acid sequences of A. thaliana and Q. suber. The proteins were named according to their gene name/EST in the Cork Oak or TAIR database (accession numbers in Table 1). The length of the branches refers to the amino acid variation rates. Numbers indicate the percentage of 1000 bootstrap re-samplings that support the inferred topology. The scale bar is an indicator of genetic distance based on branch length. (B) Relative expression of Q. suber AGP genes measured by real-time RT-PCR, and normalized to QsPP2AA3 and QsUBQ, using RNA from male or female flower tissues. Error bars indicate standard deviation (s.d.). Female, cDNA pool; male, masculine cDNA pool.
To analyse whether these genes are specific to male flowers, their expression levels were evaluated by quantitative real-time reverse transcription (qRT)-PCR in male and female flowers. The qPCR analyses showed that the putative AGP-like QsAGP15, QsAGP16L1, QsAGP20L3 and QsAGP23 genes are predominantly expressed in male flowers (Fig. 5B). QsAGP16L1, QsAGP20L3 and QsAGP23 gene expression levels in male flowers were 22-, 10- and 11-fold higher than in female flowers, respectively. Furthermore, QsAGP16L2.1 has higher transcript levels in female than in male flowers, and QsAGP20L1 transcripts in male flowers are not detectable.
DISCUSSION
Distribution of pectins and AGPs during Q. suber male gametogenesis
Q. suber male gametogenesis is similar to that observed in most angiosperms and is notable for the small and roundish size of the generative cell. Pollen grains are shed as bicellular gametophytes.
Pectins, one of the major components of plant cell walls, are present with different degrees of esterification. JIM5 antibody recognizes homogalacturonans that are partially methyl-esterified epitopes and effectively binds to a sparsely methylated homogalacturonan epitope found in the walls of a diverse range of plant species (Knox et al., 1990). The epitope structure of the JIM5 antibody also recognizes unesterified homogalacturonans (Knox et al., 1990). During Q. suber pollen development this epitope is present in the cell wall of the microsporocyte but not in the microspore, where it is seen later in development near the mature pollen pores. Only after meiosis II does this epitope recognize the tapetum cell walls and in the vesicles secreted by this tissue.
According to the Complex Carbohydrate Research Center (CCRC) JIM7 was raised against homogalacturonans partially methyl-esterified epitopes and binds to a relatively heavily methyl-esterified epitope found in cell walls of diverse species (Willats et al., 2000). In Lolium perenne (Wisniewska and Majewska-Sawka, 2006) the highly methyl-esterified pectins labelled by JIM7 can be found localized to all cells within the mature anther, including the pollen grain. Bárany et al. (2010) showed that in Capsicum annum, highly esterified pectins were good markers for proliferative cells, whereas high levels of non-esterified pectins were abundant in walls of differentiating cells. These observations suggest the involvement of pectins as indicators for cellular proliferation and differentiation during pollen development. During Q. suber pollen development this epitope is ubiquitously present in all cell walls with only small variations in labelling intensity.
LM5 recognizes RG-I pectins and in Q. suber this antibody shows intense labelling at the beginning of anther development, absent only from the tapetum and middle layer cells of the pre-meiotic anther. The labelling fades in the locule as the tetrads form but at the same time signal appears in the tapetum cell walls as well as in its secretions. In Lolium perenne (Wisniewska and Majewska-Sawka, 2006), RG-I epitopes identified with LM5 were detected only at the anther wall and they were not detected in the gametophytic cell line. LM6 recognizes α-(1,5)-L-arabinan common to both AGPs and RG-I epitopes (Willats et al., 1998). This antibody signal is similar to LM5, with a slight difference in labelling intensity.
JIM8 is an antibody that specifically detects AGP epitopes (Pennell et al., 1991). In Q. suber JIM8 labelling is very sparse on the anther cell walls but a few granules can be found along the intine and median layer. However, the tapetum and the gametophytic cell line are heavily labelled at all stages. As in arabidopsis, this antibody strongly labels the cell wall of the generative cell almost as strongly as the intine of the vegetative cell (Table 2) (Coimbra et al., 2007).
Table 2.
Comparison of the qualitative labelling intensity of the different anti-AGP antibodies used at different stages of pollen development in Arabidopsis thaliana (Coimbra et al., 2007) and Quercus suber
| Antibody |
Arabidopsis thaliana |
Quercus suber |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Microsporocytes | Tetrads | Young microspore and pollen grain |
Tapetum | Microsporocytes | Tetrads | Young microspore and pollen grain |
Tapetum | |||
| Germ line | Vesicles | Germ line | Vesicles | |||||||
| JIM8 | ++ | ++ | +++ | – | ++ | + | ++ | + | + | + |
| JIM13 | ++ | ++ | +++ | – | ++ | + | ++ | + | + | + |
| MAC207 | + | + | – | – | + | – | – | – | +++ | – |
| LM2 | + | + | – | – | + | n.d. | n.d. | n.d. | n.d. | n.d. |
The intensity of labelling is from − to +++; n.d., not determined.
JIM13 was raised against an AGP epitope (Yates et al., 1996). The labelling in cork oak is practically indistinguishable from that observed for JIM8. Another specific anti-AGP antibody (MAC207) was detected during the early stages of meiosis associated with starch granules and disappears completely at the later stages of pollen maturation. Castro and Clement (2007) suggested that starch accumulates in different cell layers of the Lilium anther during pollen maturation, and that the locule acts as a temporary sugar reservoir. This might be the case for Q. suber, suggesting that this could be a conserved mechanism.
Putative AGP-like genes in Q. suber related to male gametogenesis
In the present study, six arabidopsis AGP-like genes were identified in the Cork Oak database. All six genes encoded peptides of 63–78 amino acids in length which contained a protein backbone rich in AP, PA or SP repeats, in a range of 10–22 amino acid residues. Also, all the putative AGP peptides had a predicted N-terminal signal that is removed from the mature protein during processing and a typical C-terminal hydrophobic domain containing a GPI anchor signal sequence.
In the arabidopsis genome, 85 AGP genes were identified, including 16 AG peptides and 23 classical AGPs (Showalter et al., 2010). Recent studies on arabidopsis pollen grains and pollen tube microarray data analysis have revealed that at least 11 AGP genes were differentially expressed during pollen development (Wang et al., 2008; Qin et al., 2009; Costa et al., 2013b). Among these genes, five are highly expressed in pollen, two classical AGPs, AtAGP6 and AtAGP11, and three AG peptides, AtAGP23, AtAGP40 and AtAGP24. The AG peptides AtAGP15, AtAGP16, AtAGP20, AtAGP22 and AtAGP21 and the classical AGP1 gene also showed differential expression levels during pollen development (reviewed by Nguema-Ona et al., 2012). Additionally, functional studies using mutant analysis revealed that AtAGP11 and AtAGP6 are associated with pollen grain development and pollen tube growth (Levitin et al., 2008; Coimbra et al., 2010). By using genetics assays, AtAGP24 was suggested to be involved in non-enzymatic cleavage of the cell wall (Stenvik et al., 2006), and other functional studies demonstrated that AtAGP18 is specifically expressed in the female reproductive tissues, mainly in cell types that are involved in establishing the sporophytic to gametophytic transitions (Acosta-Garcia and Vielle-Calzada, 2004).
In this paper, we report two putative Q. suber AG peptides, QsAGP15 and QsAGP23, which shared more than 55 % deduced amino acid sequence identity with the corresponding arabidopsis orthologues. The phylogenetic tree inferred from AGP sequences showed the close relationship between these two Quercus suber AGPs and the AtAGPs highly and preferentially expressed in pollen. Furthermore, the same phylogenetic tree indicated that QsAGP15 and QsAGP23 are more closely related to the putative arabidopsis orthologues than to the other Quercus putative AGP-like genes. Although AtAGP15, in a transcriptome analysis of arabidopsis floral tissues, was reported to be down-regulated in pollen and less expressed in anthers than in carpels (Kram et al., 2009), of the Quercus putative orthologous AGPs identified here, QsAGP15 has a higher level of expression in male flowers than in female flowers.
QsAGP23, the putative orthologue of AtAGP23, is also preferentially expressed in male flowers as we have shown by qRT-PCR experiments. In previous arabidopsis pollen transcriptome analysis, AtAGP23 was identified as the AG peptide selectively expressed in pollen (Becker et al., 2003). In the arabidopsis eFP Browser (Winter et al., 2007) the AtAGP23 gene is one of AGPs expressed at very high levels during microgametogenesis. Moreover, AtAGP23 is one of five AGPs that are pollen-specific in arabidopsis (Wang et al., 2008). Curiously, the other arabidopsis pollen-specific genes, namely AtAGP6, AtAGP11, AtAGP24 and AtAGP40, have no phylogenetically related peptide sequences in the Q. suber transcriptome. As far as we know, in other angiosperms no orthologues of these arabidopsis pollen-specific genes have been found, even in rice, where several AGP-encoding genes are expressed in anthers (Ma and Zhao, 2010). However, orthologues of the pollen-specific AtAGP23 gene have been characterized in different angiosperms. In apple flowers three putative AGP-encoding genes that were specifically expressed in anthers were isolated (Choi et al., 2010) and although these genes were more closely related to apple genes than to any other arabidopsis AGP gene, they showed high homology with AtAGP23 and with BAN102, the latter a pollen-preferential gene from Chinese cabbage (Park et al., 2005). In rice, an OsAGP preferentially expressed in the inflorescence and with the deduced gene and protein sequence showing homology with the AGP-encoding gene AtAGP23 was isolated (Anand and Tyagi, 2010).
From a BLAST search in Uniprot against QsAGP23, we found peptides from tree species Prunus persica (peach) (accession no. M5X463_PRUPE) and Populus trichocarpa (Western balsam poplar) (accession no. B9HNM7_POPTR) that revealed high score similarity and more than 65 % identity with the AGP23-encoding gene from cork oak. It is remarkable that all these AGPs preferentially expressed in pollen similar to the AtAGP23, but from so distantly related Angiosperms, exhibit high homology not only in the core protein but also within the N- and C-terminal characteristic domains. These findings suggest that these AGP peptides may play similar roles during pollen development in Angiosperms.
Our results also revealed two other AG peptide-encoding genes, QsAGP16L1 and QsAGP20L3, which had much higher expression level in the male than in the female flowers. The AGP phylogenetic tree obtained revealed that these two Quercus AG peptides are the closest related among all Quercus AG peptides. In the same way, the arabidopsis AtAGP16 and AtAGP20 are the two closest AG peptides. Although these two AtAGP genes present a moderate level of expression in all plant tissues (data retrieved from the eFP browser), AtAGP16 is up-regulated during microgametogenesis preferentially in the uninucleated pollen but down-regulated during pollen germination, and AtAGP20 has a similar expression profile, despite being preferentially expressed in the bicellular pollen. In the Cork Oak database, different putative QsAGP20-like and QsAGP16-like peptide sequence homologues were found, but we have evidence at just the transcript level for the presence of two AtAGP16 and two AtAGP20 homologues in reproductive tissues. In the TAIR database, AtAGP16 appears with two variant forms, AtAGP16L1 and AtAGP16L2.1, which arise by alternative splicing. AtAGP16L2 codes for a 57 amino acid protein, 16 residues fewer than the other variant encoded transcript, and it does not have any potential GPI modification site. Similarly to arabidopsis, the QsAGP16L2 encoding gene has two splice variant forms, which differ in the size and the presence of a GPI anchor addition sequence. The longer transcript QsAGP16L2.1, which has the GPI signal sequence, codes for a 63 amino acid protein having 20 amino acids more than the shorter variant QsAGP16L2.2. In contrast to what would be expected for an AG peptide 16, the longer transcript QsAGP16L2.1 is more abundant in female flowers. These findings reveal that these AGP16 genes from two distantly related species each express two variant transcripts and that they have conserved structural features at the gene level.
These multiple genes encoding AG peptide-like 16 and/or 20 appear to have evolved from a series of gene duplications, and each duplicate member may function in different tissues, or even at diverse developmental stages. In support of this, namely that each duplicate gene may be tissue-specific, our findings showed differential transcript abundance in female and male flowers for the QsAGPs20-Like and QsAGPs16-Like genes. Moreover, the QsAGP20L1 gene, which is expressed only in female flowers, probably has a specific role in female gametogenesis. Thus, each member of these putative duplicated genes may function primarily in male or female gametogenesis.
SUPPLEMENTARY DATA
Supplementary data are available online at www.aob.oxfordjournals.org and consist of the following. Figure S1: Quercus suber predicted AG peptide sequences. Table S1: Gene-specific primers used in qRT-PCR analysis.
ACKNOWLEDGEMENTS
This work was funded by FEDER funds through the Operational Competitiveness Programme – COMPETE and by National Funds through FCT – Fundação para a Ciência e a Tecnologia under the project FCOMP-01-0124-FEDER-019461 (PTDC/AGR-GPL/118508/2010). R.S. was supported by funding from FCT with a PhD grant (ref. SFRH/BD/84365/2012).
LITERATURE CITED
- Acosta-Garcia G, Vielle-Calzada JP. 2004. A classical arabinogalactan protein is essential for the initiation of female gametogenesis in arabidopsis. Plant Cell 16: 2614–2628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anand S, Tyagi AK. 2010. Characterization of a pollen-preferential gene OSIAGP from rice (Oryza sativa L. subspecies indica) coding for an arabinogalactan protein homologue, and analysis of its promoter activity during pollen development and pollen tube growth. Transgenic Research 19: 385–397. [DOI] [PubMed] [Google Scholar]
- Azevedo H, Lino-Neto T, Tavares RM. 2003. An improved method for high-quality RNA isolation from needles of adult maritime pine trees. Plant Molecular Biology Reporter 21: 333–338. [Google Scholar]
- Bárany IL, Fadón B, Risue-Risueno MC, Testillano PS. 2010. Cell wall components and pectin esterification levels as markers of proliferation and differentiation events during pollen development and pollen embryogenesis in Capsicum annum L. Journal of Experimental Botany 61: 1159–1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker JD, Boavida LC, Carneiro J, Haury M, Feijó JA. 2003. Transcriptional profiling of arabidopsis tissues reveals the unique characteristics of the pollen transcriptome. Plant Physiology 133: 713–725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boavida LC, Varela MC, Feijó J. 1999. Sexual reproduction in the cork oak (Quercus suber L.). I. The progamic phase. Sexual Plant Reproduction 11: 34711:3 [Google Scholar]
- Boavida LC, Silva JP, Feijó S J. 2001. Sexual reproduction in the cork oak (Quercus suber L). II. Crossing intra- and interspecific barriers. Sexual Plant Reproduction 14: 14314:1. [Google Scholar]
- Castro AJ, Clement C. 2007. Sucrose and starch catabolism in the anther of Lilium during its development: a comparative study among the anther wall, locular fluid and microspore/pollen fractions. Planta 225: 1573–1582. [DOI] [PubMed] [Google Scholar]
- Chang S, Puryear J, Cainey J. 1993. Simple and efficient method for isolating RNA from pine trees. Plant Molecular Biology Reports 11: 113–116. [Google Scholar]
- Choi YO, Kim SS, Lee S, et al. 2010. Isolation and promoter analysis of anther-specific genes encoding putative arabinogalactan proteins in Malus×domestica. Plant Cell Reports 29: 15–24. [DOI] [PubMed] [Google Scholar]
- Coimbra S, Almeida J, Junqueira V, Costa ML, Pereira LG. 2007. Arabinogalactan proteins as molecular markers in Arabidopsis thaliana sexual reproduction. Journal of Experimental Botany 58: 4027–4035. [DOI] [PubMed] [Google Scholar]
- Coimbra S, Costa M, Jones B, Mendes MA, Pereira LG. 2009. Pollen grain development is compromised in arabidopsis agp6 agp11 null mutants. Journal of Experimental Botany 60: 3133–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coimbra S, Costa M, Mendes MA, et al. 2010. Early germination of arabidopsis pollen in a double null mutant for the arabinogalactan protein genes AGP6 and AGP11. Sexual Plant Reproduction 23: 199–205. [DOI] [PubMed] [Google Scholar]
- Costa M, Pereira AM, Rudall PJ, Coimbra S. 2013a. Immunolocalization of arabinogalactan proteins (AGPs) in reproductive structures of an early-divergent angiosperm, Trithuria (Hydatellaceae). Annals of Botany 111: 183–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa M, Nobre MS, Becker JD, et al. 2013b. Expression-based and co-localization detection of arabinogalactan protein 6 and arabinogalactan protein 11 interactors in arabidopsis pollen and pollen tubes. BMC Plant Biology 8: 13:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenhaber B, Wildpaner M, Schultz CJ, et al. 2003. Glycosylphosphatidylinositol lipid anchoring of plant proteins. Sensitive prediction from sequence- and genome-wide studies for arabidopsis and rice. Plant Physiology 133: 1691–1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farmer RE., Jr 1981. Variation in seed yield of white oak. Forest Science 27: 377–380. [Google Scholar]
- Knox JP, Linstead PJ, King J, Cooper C, Roberts K. 1990. Pectin esterification is spatially regulated both within cell walls and between developing tissues of root apices. Planta 181: 512–521. [DOI] [PubMed] [Google Scholar]
- Knox JP, Linstead PJ, Peart J, Cooper C, Roberts K. 1991. Developmentally-regulated epitopes of cell surface arabinogalactan-proteins and their relation to root tissue pattern formation. Plant Journal 1: 317–326. [DOI] [PubMed] [Google Scholar]
- Johnson KL, Jones BJ, Bacic A, Schultz CJ. 2003. The fasciclin-like arabinogalactan proteins of arabidopsis. A multigene family of putative cell adhesion molecules. Plant Physiology 133: 1911–1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones L, Seymour GB, Knox JP. 1997. Localization of pectic galactan in tomato cell walls using a monoclonal antibody specific to (1→4)-β-d-galactan. Plant Physiology 113: 1405–1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kram BW, Xu WW, Carter CJ. 2009. Uncovering the Arabidopsis thaliana nectary transcriptome: investigation of differential gene expression in floral nectariferous tissues. BMC Plant Biology 15: 9:92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levitin B, Richter D, Markovich I, Zik M. 2008. Arabinogalactan proteins 6 and 11 are required for stamen and pollen function in arabidopsis. Plant Journal 56: 351–363. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔC(T) method . Methods 25: 402–408. [DOI] [PubMed] [Google Scholar]
- Ma H, Sundaresan V. 2010. Development of flowering plant gametophytes. Current Topics in Developmental Biology 91: 379–412. [DOI] [PubMed] [Google Scholar]
- Ma H, Zhao J. 2010. Genome-wide identification, classification, and expression analysis of the arabinogalactan protein gene family in rice (Oryza sativa L.). Journal of Experimental Botany 61: 2647–2668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguema-Ona E, Coimbra S, Vicré M, Mollet JC, Driouich A. 2012. Arabinogalactan proteins in root and pollen-tube cells: distribution and functional aspects. Annals of Botany 110: 383–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park BS, Kim JS, Kim SH, Park YD. 2005. Characterization of a pollen-preferential gene, BAN102, from Chinese cabbage. Plant Cell Reports 24: 663–670. [DOI] [PubMed] [Google Scholar]
- Pennell RI, Knox JP, Scofield GN, Selvendran R, Roberts K. 1989. A family of abundant plasma membrane-associated glycoproteins related to the arabinogalactan proteins is unique to flowering plants. Journal of Cell Biology 108: 1967–1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennell RI, Janniche L, Kjellbom P, Scofield GN, Peart JM, Roberts K. 1991. Developmental regulation of a plasma membrane arabinogalactan protein epitope in oilseed rape flowers. Plant Cell 3: 1317–1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira-Leal JB, Abreu IA, Alabaça CS, et al. 2014. A comprehensive assessment of the transcriptome of cork oak (Quercus suber) through EST sequencing. BMC Genomics 15: 15:371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen T, Brunak S, von Heijne G, Nielsen H. 2011. Signal P 4.0: discriminating signal peptides from transmembrane regions. Nature Methods 8: 785–786. [DOI] [PubMed] [Google Scholar]
- Qin Y, Leydon AR, Manziello A, et al. 2009. Penetration of the stigma and style elicits a novel transcriptome in pollen tubes, pointing to genes critical for growth in a pistil. PLoS Genetics 5: e1000621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz CJ, Rumsewicz MP, Johnson KL, et al. 2002. Using genomic resources to guide research directions. The arabinogalactan protein gene family as a test case. Plant Physiology 129: 1448–1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seifert G, Roberts K. 2007. The biology of arabinogalactan proteins. Annual Review of Plant Biology 58: 137–161. [DOI] [PubMed] [Google Scholar]
- Showalter AM, Keppler B, Lichtenberg J, Gu D, Welch LR. 2010. A bioinformatics approach to the identification, classification, and analysis of hydroxyproline-rich glycoproteins. Plant Physiology 153: 485–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenvik GE, Butenko MA, Urbanowicz BR, Rose JK, Aalen RB. 2006. Overexpression of INFLORESCENCE DEFICIENT IN ABSCISSION activates cell separation in vestigial abscission zones in arabidopsis. Plant Cell 18: 1467–1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephen F, Altschul JC, Wootton E, et al. 2005. Protein database searches using compositionally adjusted substitution matrices. FEBS Journal 272: 5101–5109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Molecular Biology Evolution 28: 2731–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson T. 1994. Clustal W improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Research 22: 4673c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twell D. 2011. Male gametogenesis and germline specification in flowering plants. Sexual Plant Reproduction 24: 149–160. [DOI] [PubMed] [Google Scholar]
- Varela MC, Valdiviesso T. 1996. Phenological phases of Quercus suber L. flowering. Forest Genetics 3: 93–102. [Google Scholar]
- Willats WGT, Marcus SE, Knox JP. 1998. Generation of a monoclonal antibody specific to (1→5)-α-l-arabinan. Carbohydrate Research 308: 149–152. [DOI] [PubMed] [Google Scholar]
- Willats WGT, Limberg G, Buchholt HC, et al. 2000. Analysis of pectic epitopes recognised by hybridoma and phage display monoclonal antibodies using defined oligosaccharides, polysaccharides, and enzymatic degradation. Carbohydrate Research 327: 309–320. [DOI] [PubMed] [Google Scholar]
- Wang Y, Zhang WZ, Song LF, et al. 2008. Transcriptome analyses show changes in gene expression to accompany pollen germination and tube growth in arabidopsis. Plant Physiology 148: 1201–1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter D, Vinegar B, Nahal H, Ammar R, Wilson GV, Provart NJ. 2007. An Electronic Fluorescent Pictograph browser for exploring and analyzing large-scale biological data sets. PLoS One 2: e718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wisniewska E, Majewska-Sawka A. 2006. Cell wall polysaccharides in differentiating anthers and pistils of Lolium perenne. Protoplasma 228: 65–71. [DOI] [PubMed] [Google Scholar]
- Yates EA, Valdor J-F, Haslam SM, et al. 1996. Characterization of carbohydrate structural features recognized by anti-arabinogalactan-protein monoclonal antibodies. Glycobiology 6: 131–139. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.