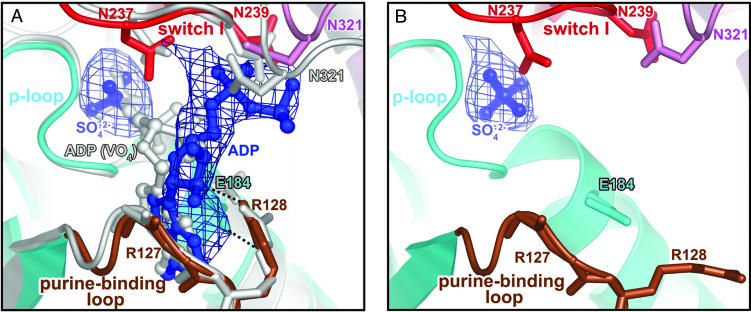
Fig. 1.
Overview of the nucleotide-binding pocket. (A) ADP (blue) in the scallop myosin near-rigor structure adopts a partially bound conformation at the front door of the nucleotide-binding pocket. For comparison, the fully bound ADP from the S1–ADP·VO4 complex in the scallop prepower-stroke structure (PDB ID code 1QVI) is shown in light gray. Also shown is an Fo – Fc simulated-annealing electron density map at a 1.0 σ contour level, calculated after omitting ADP and sulfate. Residues in switch I (red) and N321 (pink) from the 50-kDa upper subdomain that are involved in weak interactions (see text) with the ADP diphosphate moiety are shown in stick representation. The same residues from the prepower-stroke conformation structure are shown in light gray. The only major difference between the two structures in the nucleotide-binding pocket of the enzyme is that N321 (50-kDa upper subdomain) makes closer contact with the bound nucleotide in the prepower-stroke structure. The side chain of R127 and the hydrogen bond between its main-chain amide and the ADP ribose O2′ are not shown for clarity. The P-loop and purine-binding loop are shown in cyan and brown, respectively. E184 (cyan sticks) and R128 (brown sticks) in the two loops form a salt bridge (dashed lines) that protects one face of the ADP base from solvent. (B) Corresponding view of the ScS1–SO4 structure, with an Fo – Fc simulated-annealing electron density map at a 2.0 σ contour level, calculated after omitting the sulfate. The E184/R128 salt bridge is not formed in the absence of a nucleotide, supporting the role of R128 in nucleotide recruitment.