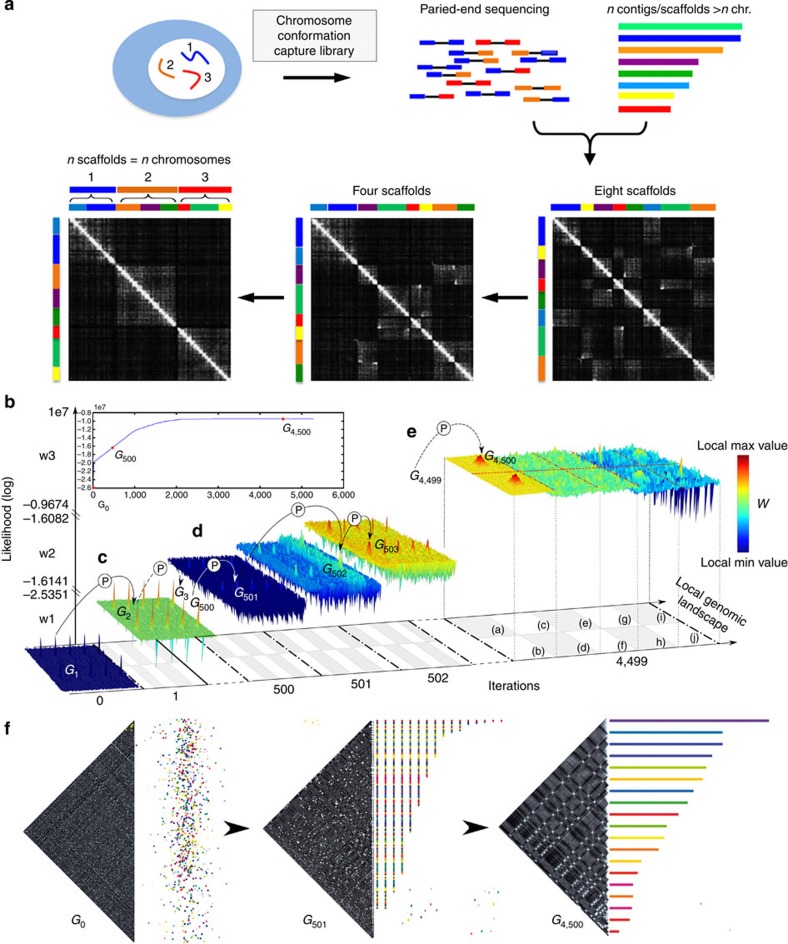
Figure 1. Principle of genomic assembly from chromosomal contact data using GRAAL.
(a) Upper panels, from left to right: a fictitious genome comprising three chromosomes is processed into a genomic Hi-C library and then paired-end-sequenced. The fictitious genome is not fully assembled, but remains split into eight contigs or scaffolds. To resolve the genome structure, the reads from the Hi-C library are mapped on these contigs, allowing the construction of an initial contact matrix. Lower panels, from right to left: the presence of off-diagonal blocks in this initial contact matrix reflects the imperfections in the original assembly. GRAAL iteratively modifies the set of contigs in order to remove progressively these features and to increase the likelihood of the genome structure given the Hi–C data. In the final steps, the off-diagonal blocks disappear and genome structures that better reflect the 3D contact data are recovered. (b) Detailed visualization of the sampling algorithm implemented in GRAAL at three different stages: (c) initialization (iterations 0 and 1); (d) rapid increase in likelihood (iterations 500–502); (e) stabilization and fine-tuning of the structure (iteration 4,500). Because of the huge jumps in likelihood space performed by GRAAL, different scales are used for each of the three windows represented in c–e. The likelihoods on the z axis are represented using the same colour scale for windows w1, w2 and w3 (right panel). Hi-C reads are aligned on a reference genome G and the algorithm is initialized with G0, the set of contigs obtained by splitting G into bins of two or more restriction fragments (as determined by the user). At each iteration, a bin is picked at random. This bin is used to explore the local genomic landscape of structural variations around the current genomic structure, whose distribution is represented along the x and y axes. The planes occupied by the different structural variants are detailed in e: single insertion (a, b), insertion and split of contigs (c–f) and translocations (g–j). On the basis of the likelihoods computed for these structures (z axis), the sampling algorithm selects the next genomic structure, and a new set of nuisance parameters is sampled (white circles with the letter P, see methods). The algorithm reaches an equilibrium after ~3,000 iterations, which corresponds to the target distribution of optimum genome structures as displayed in b. (f) Real-time visualization of both the new scaffolds (left) and the corresponding contact maps (right) allows visual monitoring of the progress of the assembly (see Supplementary Movie 1).