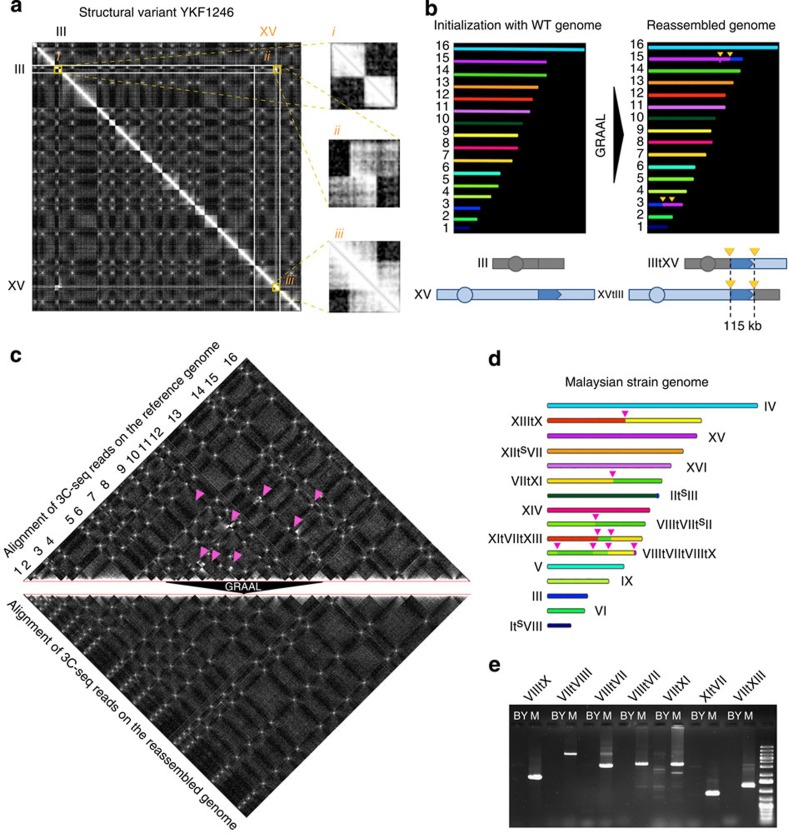
Figure 3. Identification of structural genome rearrangements using GRAAL.
(a) S. cerevisiae strain YKF1246 contact matrix obtained by mapping 3C-seq data to the reference wild-type genome revealed intra- (i, iii) and interchromosomal (ii) incongruities affecting chromosomes 3 and 15. (b) GRAAL initialized with the reference genome and the YKF1246 contact matrix generated a reassembled genome carrying a ~100-kb duplication and a translocation between chromosomes 3 and 15 (yellow arrows), as expected22. (c) Malaysian yeast strain UWOPS03-461.4 contact matrix obtained by mapping 3C-seq data to the reference wild-type S. cerevisiae genome (top panel) revealed interchromosomal incongruities (pink arrows). GRAAL reconstructed a genome structure exempt of these features (bottom panel). (d) Structure of the Malaysian yeast strain, with pink arrows representing the breakpoints of the translocations readily visible in the contact matrix of c, top. (e) Experimental PCR validation of the breakpoints identified by GRAAL. BY: reference strain (BY4741). M: Malaysian strain.