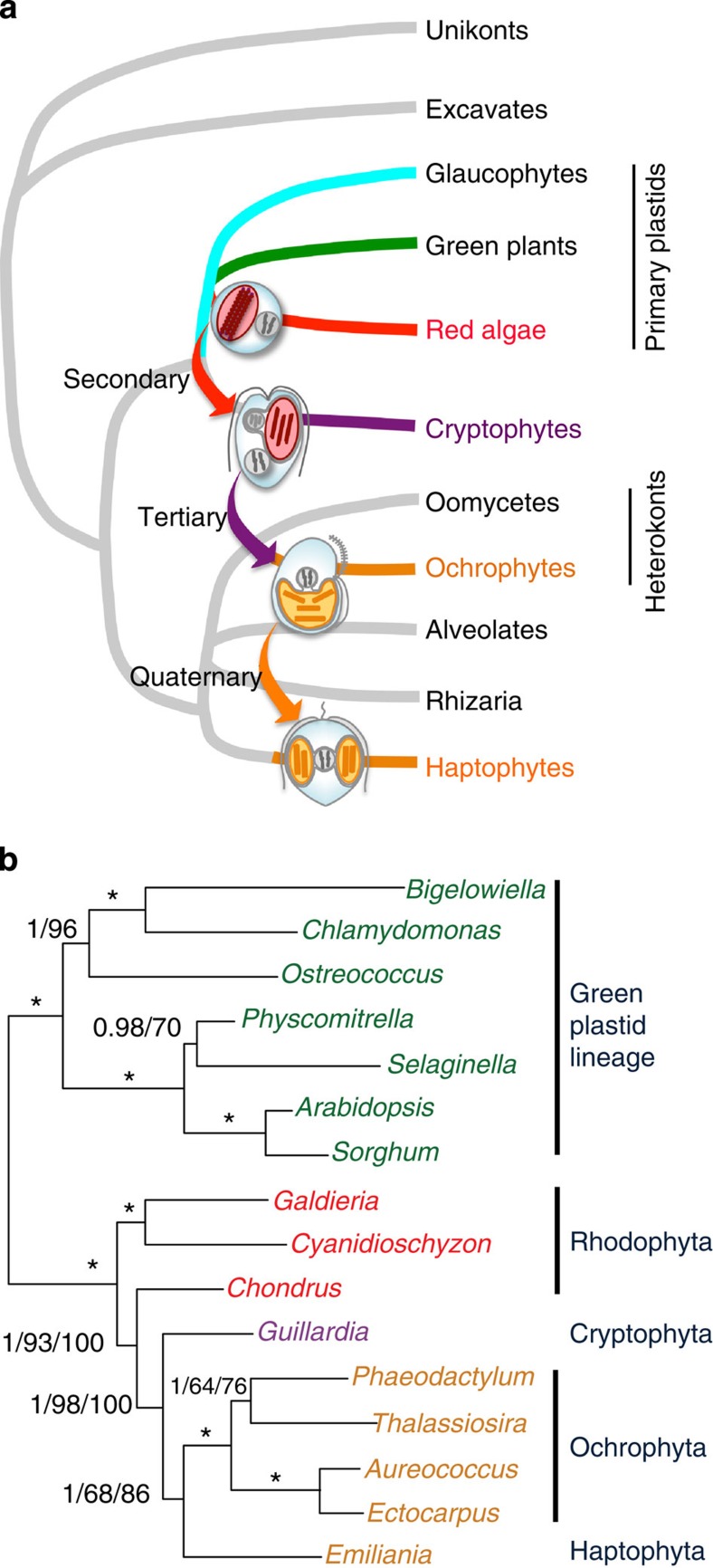
Figure 2. Model of serial plastid endosymbioses and a test using genes from plastid genomes.
(a) Model of serial plastid endosymbioses suggested by regression analyses. The relationships depicted agree with general inferences from eukaryotic phylogenomics that heterokonts are not closely related to cryptophytes or haptophytes, and that each of the three groups emerge from mutually exclusive clades containing heterotrophic relatives. With respect to our proposed model of serial endosymbioses, the specific topology of the tree is not important, only that the three chromist algal groups do not form an exclusively monophyletic grouping that excludes aplastidial heterokonts. (b) Tree of plastid relationships based on an alignment of 5,818 amino-acid positions from genes inherited directly through the plastid genome, an independent data set for testing the model of plastid transfer inferred from EGT. The tree shown was recovered using both Bayesian and maximum-likelihood (ML) approaches. Bayesian posterior probabilities and ML bootstrap support values are provided for each node, with a star indicating 1.0 and 100% support, respectively. A second bootstrap value on nodes in the red plastid clade is from ML analyses performed in the absence of the green plastid lineage as outgroup. Bayesian probabilities are based on 5,000 sampled trees and bootstrap support values are from 1,000 replicates in each case.