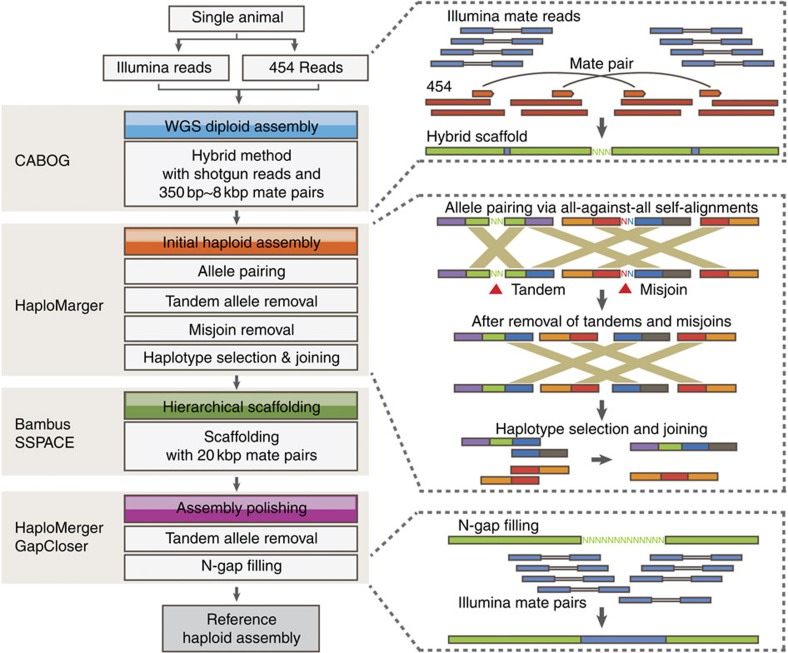
Figure 1. A novel whole-genome shotgun (WGS) assembly pipeline for highly polymorphic diploid genomes.
The pipeline was gradually set-up to achieve optimal assembly quality through testing and combining algorithms and data sets. An upgraded version of HaploMerger7 was used to monitor assembly quality, to correct major assembly errors such as misjoins and tandem misassemblies and to separate and reconstruct haploid assemblies. We chose the assembler CABOG44 for de novo hybrid assembly to compensate for the short-read lengths and different sequencing error types by combining the advantages of 454 reads and Illumina reads. We conducted further hierarchical scaffolding of pre-assembled contigs using SSPACE45. GapCloser46 was employed to close N-gaps. Details of the pipeline and its development, application and assessment are described in Supplementary Note 2.