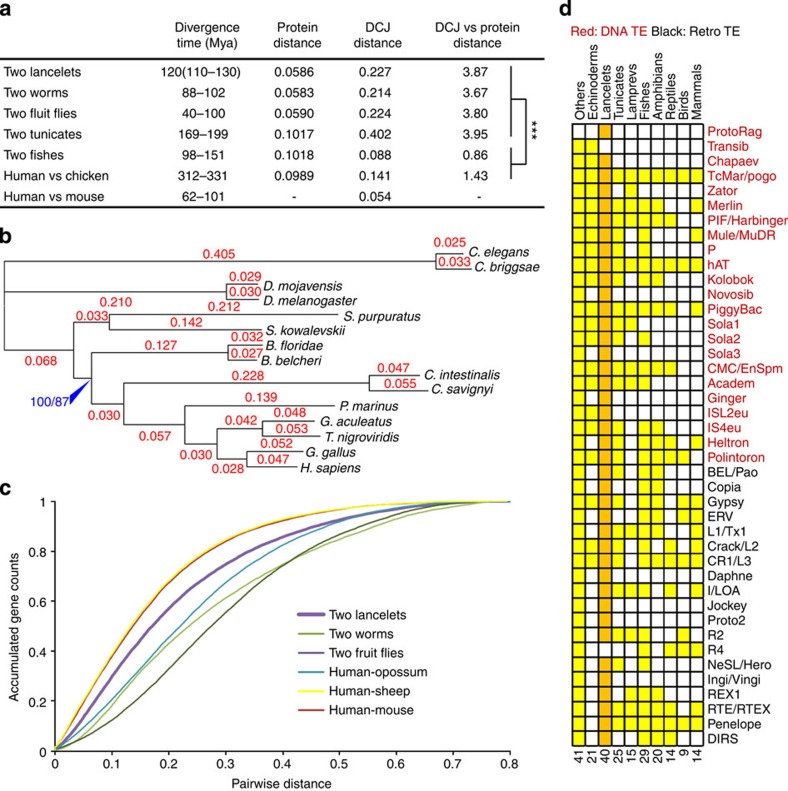
Figure 2. Comparative analysis of molecular divergence and TEs.
(a) Comparison of divergence times of selected species pairs (see Supplementary Table 4 and Supplementary Note 3 for the source of divergence times), protein distances (based on the conserved amino-acid sites of 729 orthologous genes present in 15 widely divergent species), DCJ distances (based on all orthologous protein genes of the species pair) and relative DCJ distances (DCJ distance divided by protein distance). *** indicates significant difference (P<1e−16, χ2-test). (b) Maximum-likelihood (ML) phylogenetic tree containing the numbers of expected substitutions per amino-acid position, using 245,205 conserved sites from a concatenated alignment of 729 orthologous protein genes. Both Bayesian supports and ML bootstrap supports were 100% for all nodes but one, whose statistical support (Bayesian/ML) is indicated in blue colour. Supplementary Fig. 3 and Supplementary Note 3 provide details of this phylogenetic analysis. (c) The cumulative distribution of the pairwise protein distances of all 1:1 orthologues in the six species pairs. Note that the curve of human versus mouse largely overlaps with that of human versus sheep. The orthologous protein distance between the two lancelet species falls midway between those of human versus sheep (divergence time: 95–113 Myr) and human versus opossum (divergence time: 125–138 Myr). More information is provided in Supplementary Note 3. (d) Distribution of the ATE superfamilies in the major animal lineages. For lancelets, ATE families are required to be present in both Florida and Chinese lancelets; for the other lineages, TE families are required to be present in at least one species of that lineage. Data for other lineages were taken from RepBase and the literature. More information is provided in Supplementary Note 6.