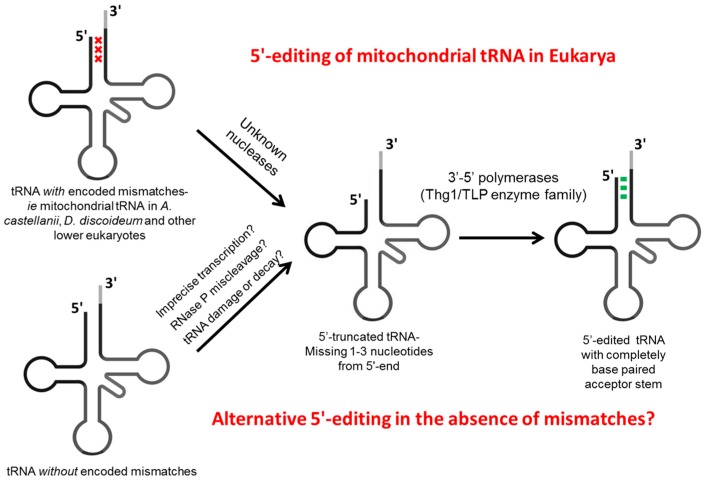
Figure 2.
Known and predicted 5'-end editing pathways. 5'-end editing of mt-tRNA is associated with the presence of mismatched nucleotides at any of the first three positions of the tRNA acceptor stem (indicated by the red X). The top pathway shows the two-step editing process that repairs these mismatches, first by removal of the 5'-mismatched residues (by unknown enzymes) and next by repair of the resulting 5'-truncated tRNA by 3'–5'-polymerases known as TLPs (Thg1-like proteins; Thg1: tRNAHis guanylyltransferase), thus generating a fully base paired acceptor stem (Watson–Crick base pairs indicated by the green bars). The bottom pathway shows an alternative process that could also utilize similar 3'–5'-polymerases to edit tRNA in the absence of bona fide mismatches, such as in organisms where these types of mismatched nucleotides have not been identified in tRNA genes. In these cases, several possible pathways, as indicated, could conceivably generate 5'-truncated substrates to be repaired by Thg1/TLP orthologs found in these species.