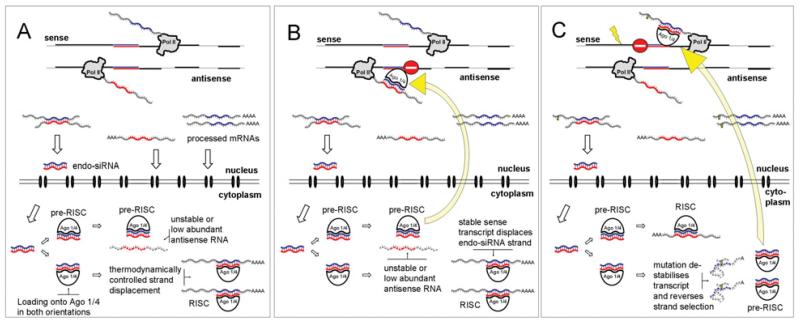
Figure 2. Working model of gene regulation by NATs.
(A and B) Representation of the physiological situation where a stable, correctly folded, protein-coding sense transcript is produced. Upon transcription and processing of both sense and antisense transcripts, the RNAs are either processed into endo-siRNAs or transported into the cytoplasm. In the cytoplasm, the endo-siRNAs are loaded on to a non-slicing Argonaute protein (most probably Ago4 or Ago1) in both orientations (pre-RISC). The availability of a target sequence will drive RISC formation [60]. The resulting complex will stay in the cytoplasm and the mRNA may eventually become translated. If no target RNA molecules are available, the pre-RISC complex is transported back into the nucleus and induces silencing of the complementary transcript. (C) Situation where a mutation changes the stability or the folding of the protein-coding sense transcript. As a consequence, RISC is not formed with the sense but with the now more abundant antisense RNA, and the mutated sense transcript will become silenced. We can only speculate on the allele-specificity of the process; however, it is likely that the timing of the entire process, transcription→processing→feedback, is essential.