Abstract
BACKGROUND:
Relationships between airway inflammation and respiratory potentially pathogenic microorganisms (PPMs) quantified using quantitative polymerase chain reaction (qPCR) in subjects with COPD are unclear. Our aim was to evaluate mediators of airway inflammation and their association with PPMs in subjects with COPD at stable state and during exacerbations.
METHODS:
Sputum from 120 stable subjects with COPD was analyzed for bacteriology (colony-forming units; total 16S; and qPCR targeting Haemophilus influenzae, Moraxella catarrhalis, and Streptococcus pneumoniae), differential cell counts, and inflammatory mediators using the Meso-Scale Discovery Platform. Subjects were classified as colonized if any PPM was identified above the threshold of detection by qPCR. Symptoms were quantified using the visual analog scale.
RESULTS:
At stable state, 60% of subjects were qPCR positive for H influenzae, 48% for M catarrhalis, and 28% for S pneumoniae. Elevated sputum concentrations of IL-1β, IL-10, and tumor necrosis factor (TNF)-α were detected in samples qPCR positive for either H influenzae or M catarrhalis. Bacterial loads of H influenzae positively correlated with IL-1β, IL-8, IL-10, TNF-α, and symptoms; and M catarrhalis correlated with IL-10 and TNF-α. H influenzae qPCR bacterial load was an independent predictor of sputum TNF-α and IL-1β. In 55 subjects with paired exacerbation data, qPCR bacterial load fold change at exacerbation in M catarrhalis but not H influenzae correlated to changes in sputum TNF-α and IL-1β concentrations.
CONCLUSIONS:
At stable state, H influenzae is associated with increased airway inflammation in COPD. The relationship between bacterial load changes of specific pathogens and airway inflammation at exacerbation and recovery warrants further investigation.
COPD is characterized by irreversible airflow obstruction and airway inflammation. The disease course is punctuated by exacerbation episodes,1 which are often associated with increased airway inflammation,2 viruses, and bacteria.3 Bacteria are isolated from sputum cultures in 30% to 40% of subjects with stable COPD3‐7 and found in approximately 50% of subjects during exacerbation episodes.7,8 Colonization with bacteria is associated with worsened health status,6 reduced lung function,9 and an increase in the frequency and severity of exacerbations.10 Additionally, patients with positive sputum cultures have an associated increased inflammatory response detected by elevated levels of sputum neutrophils,11 IL-8,12‐14 IL-6,13 tumor necrosis factor (TNF)-α,13 myeloperoxidase,14 and leukotriene B4.6 Both culture-based and culture-independent molecular techniques have shown that Haemophilus influenzae is the commonest sputum pathogen in stable COPD.15,16 Although studies using culture-based techniques have suggested that airway inflammation is higher in those colonized with H influenzae,15‐17 relationships between pathogens quantified using molecular techniques and airway inflammation are unclear. In this study using quantitative polymerase chain reaction (qPCR) to measure pathogen-specific bacterial loads, we hypothesized that detection of the respiratory potentially pathogenic microorganisms (PPMs) (H influenzae, Moraxella catarrhalis, Streptococcus pneumoniae, and Staphylococcus aureus) in subjects with COPD is associated with increased airway inflammation at stable state and during exacerbations.
Materials and Methods
Subjects
Sputum samples from subjects aged ≥ 40 years and with postbronchodilator FEV1/FVC < 0.7 enrolled within an observational COPD exacerbation cohort study were analyzed. The study design and inclusion and exclusion criteria have been described previously.3 Subjects with COPD on prophylactic antibiotic therapy were excluded. The study was conducted in accordance with the amended Declaration of Helsinki and was approved by the Leicestershire, Northamptonshire, and Rutland ethics committee (07/H0406/157). All patients gave informed written consent.
Measurements
Baseline demographic information, smoking history, medication history, and patient-reported history of exacerbations were collected. Subjects were reviewed when clinically stable and during exacerbation episodes; stable visits took place a minimum of 8 weeks after exacerbation episodes. Exacerbations were defined according to the criteria of Anthonisen et al18 and health-care use19 and treated according to guidelines.20 Exacerbation testing and sampling was only performed in subjects who were treatment naive for the episode. At all visits, spirometry and symptom assessment using the St. George’s Respiratory Questionnaire (SGRQ),21 the Chronic Respiratory Questionnaire,22 and the visual analog scale (VAS)23 were undertaken. Spontaneous or induced sputum sampling was collected for analysis of microbiology, differential cell counts, and cytokine analysis, as described later. No differences in inflammatory counts between spontaneous or induced sputum samples were identified in this study, in keeping with other studies,24,25 and > 95% of subjects provided spontaneous sputum samples. CT imaging, to investigate bronchiectasis, was not performed as part of the study protocol; however, CT scans performed as part of a routine clinical investigation were interrogated in subjects who entered the study.
Sputum Assessments
Bacterial load was measured by colony-forming units (CFU) as per standard methods26 and quantitative polymerase chain reaction (qPCR) as previously described.3 The CFU is a semiquantitative analysis of total live bacterial counts, and a quantitative analysis of both live and dead bacteria was quantified using qPCR, estimating both the total bacterial load based on the abundance of 16S ribosomal subunit encoding genes (total 16S). Pathogen-specific bacterial 16S abundance using qPCR was measured for H influenzae, M catarrhalis, S pneumoniae, and S aureus (Pseudomonas aeruginosa was not measured by qPCR in this study). Quantification of the total bacterial load of H influenzae and S aureus was performed using the SYBR Green assay (Life Technologies). The TaqMan assay (Life Technologies) was used to quantify M catarrhalis and S pneumoniae (target genes and primers listed in e-Table 1 (461KB, pdf) ). S aureus was infrequently detected, and therefore any results relating to this pathogen were not analyzed further. The threshold of detection for pathogen-specific bacterial 16S qPCR analysis and CFU counts was taken as 1 × 104 genome copies/mL and 1 × 105 colonies/mL of sputum, respectively, reflecting previous cutoff thresholds used in this field.27,28 Subjects were categorized as pathogen-specific bacterial 16S qPCR positive detection if any qPCR PPM (defined in this study as identification of H influenzae, M catarrhalis, S pneumoniae, and S aureus) was identified above the threshold. Subjects were classified as codetection if more than one pathogen-specific bacterial 16S qPCR PPM was identified above the threshold of identification. Sputum was simultaneously processed to obtain cytospins for differential cell counts and cell-free supernatants as previously described.29 The sputum inflammatory mediators IL-1β, IL-5, IL-6, IL-8, IL-10, TNF-α, TNFRI, CXCL10, CCL2, CCL3, CCL4, CCL5, CCL13, and CCL17 were measured using the Meso Scale Discovery Platform (Meso Scale Diagnostics, LLC) from sputum supernatants.25 All mediators, except IL-10, measured using the MSD were detectable and above the limit of detection in > 75% of samples (IL-10 was above the limit of detection in > 50% of samples; limits of detection presented in e-Table 2 (461KB, pdf) ).
Statistical Analysis
Statistical analysis was performed using SPSS version 20 (IBM) and PRISM version 6 (GraphPad Software, Inc). Parametric data were expressed as mean (SEM), nonparametric data as median (interquartile range), and log-normally distributed data as geometric mean (95% CI). Unpaired parametric and nonparametric groups were compared using the Student t test and Mann-Whitney test, respectively. The paired t test was used to compare matched stable and exacerbation measures of airway bacterial load and sputum inflammatory mediators. For comparison of three or more groups, the one-way analysis of variance was used, with repeated analysis of variance for paired data. The χ2 test was used to compare proportions between groups. Pearson correlation coefficient was used to assess correlations between qPCR-measured airway bacterial load and sputum inflammatory mediators. Multivariate stepwise regression analysis was performed to model the effects of bacterial load on proinflammatory cytokine expression, namely sputum TNF-α and IL-1β. Variables that demonstrated significance at the P < .10 level using univariate analysis were entered into the model: H influenzae and M catarrhalis bacterial load, sputum total cell count, percentage sputum neutrophils, and CFU. Exacerbation frequency and % FEV1 predicted were also entered into the model for clinical relevance. The regression model did not show violation of multicollinearity or homoscedasticity, and the residuals observed normality. No corrections for multiple mediator measurements were performed. A P value < .05 was taken as the threshold of significance for all statistical testing.
Results
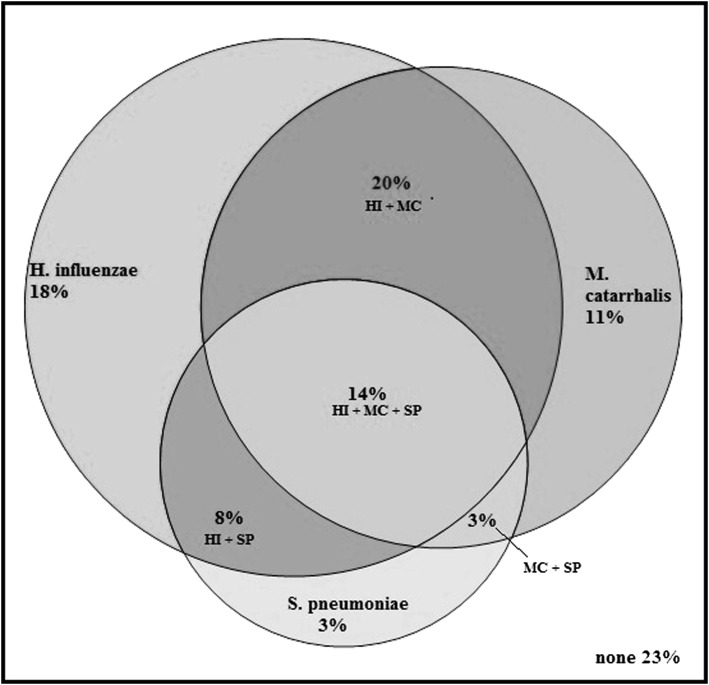
Stable sputum samples with full complement of inflammatory mediators were obtained from 120 subjects (83 men). The clinical characteristics are presented in Table 1. A CT scan was available in 93 subjects (77.5%), and bronchiectasis was detected in 18 (19.4%). There were no significant differences in the clinical parameters between the subjects with COPD with or without detectable pathogen-specific bacterial 16S qPCR PPM. Subjects with PPMs on qPCR had more severe airflow obstruction and increased CFU but not total 16S (Table 1). There was no correlation between total 16S qPCR and inhaled corticosteroid dose, smoking pack-years, or exacerbation frequency. The distribution of qPCR pathogen codetection is presented in Figure 1.
TABLE 1 ] .
Clinical Characteristics of Subjects According to Whether There Was a Pathogen-Specific Bacterial 16S qPCR PPM Detected Above the Limit of Detectiona
| Characteristic | qPCR PPM Negative (n = 28) | qPCR PPM Positive (n = 92) | P Value |
| Age, mean (range), y | 70 (48-87) | 69 (43-88) | .37 |
| Ex-smokers, No. (%) | 20 (71) | 63 (68) | .82 |
| Proportion on ICS, No. (%) | 24 (86) | 81 (88) | .75 |
| ICS dose, median (IQR), mg (BDP equivalent) | 1,300 (800-2,000) | 2,000 (800-2,000) | .55 |
| Pack-y history | 45 (10-120) | 49 (10-153) | .56 |
| FEV1, L | 1.47 (0.09) | 1.32 (0.06) | .20 |
| FEV1/FVC % | 61 (4) | 53 (2) | .05 |
| FEV1 % | 59 (4) | 51 (2) | .06 |
| Exacerbations in previous y | 3.8 (0.5) | 3.5 (0.3) | .59 |
| SGRQ symptoms, units | 62.2 (94.1) | 57.2 (2.4) | .31 |
| SGRQ activities, units | 70.2 (4.0) | 65.7 (2.3) | .35 |
| SGRQ impacts, units | 34.7 (3.6) | 37.0 (2.0) | .58 |
| SGRQ total, units | 49.6 (3.3) | 48.7 (1.9) | .82 |
| CRQ emotion, units | 4.7 (0.2) | 4.7 (0.2) | .97 |
| CRQ fatigue, units | 3.6 (0.2) | 3.6 (0.1) | .92 |
| CRQ dyspnea, units | 3.4 (0.2) | 3.3 (0.1) | .69 |
| CRQ mastery, units | 4.9 (0.3) | 4.9 (0.2) | .82 |
| CRQ total, units | 4.2 (0.2) | 4.1 (0.1) | .10 |
| VAS cough, mm | 35 (6) | 39 (3) | .45 |
| VAS dyspnea, mm | 50 (5) | 47 (3) | .64 |
| VAS sputum production, mm | 29 (5) | 38 (3) | .13 |
| VAS sputum purulence, mm | 33 (5) | 29 (3) | .55 |
| Log CFU/mL, mean (95% CI) | 6.6 (5.4-7.8) | 7.5 (5.9-9.1) | < .01 |
| Log 16S genome copies/mL, mean (95% CI) | 8.2 (6.8-9.4) | 8.4 (6.5-9.8) | .37 |
| Sputum total cell count, × 106/g | 2.5 (1.8-3.7) | 3.6 (2.5-5.1) | .27 |
| Sputum neutrophils, mean (range), % | 64 (4) | 72 (2) | .09 |
| Sputum neutrophil count, × 106/g (geometric mean, 95% CI) | 1.6 (0.9-2.8) | 2.4 (1.7-3.4) | .22 |
| Sputum eosinophils, % (geometric mean, 95% CI) | 1.6 (1.1-2.2) | 1.3 (0.9-1.9) | .61 |
| Proportion with bronchiectasis, No. (%)b | 4 of 19 (21) | 14 of 74 (18) | .74 |
Data expressed as mean (SEM) unless otherwise stated. BDP = beclomethasone dipropionate; CFU = colony-forming units; CRQ = Chronic Respiratory Questionnaire; ICS = inhaled corticosteroid; PPM = potentially pathogenic microorganism; qPCR = quantitative polymerase chain reaction; SGRQ = St. George’s Respiratory Questionnaire; VAS = visual analog scale.
> 1 × 104 genome copies/mL.
CT scans available in 93 out of 120 subjects.
Figure 1 –
Proportional Venn diagram depicting the distribution of qPCR codetection at baseline for all subjects. H. influenzae = Haemophilus influenzae; HI = Haemophilus influenzae; M. catarrhalis = Moraxella catarrhalis; MC = Moraxella catarrhalis; qPCR = quantitative polymerase chain reaction; S. pneumoniae = Streptococcus pneumoniae; SP = Streptococcus pneumoniae.
qPCR and Inflammation During Stable State
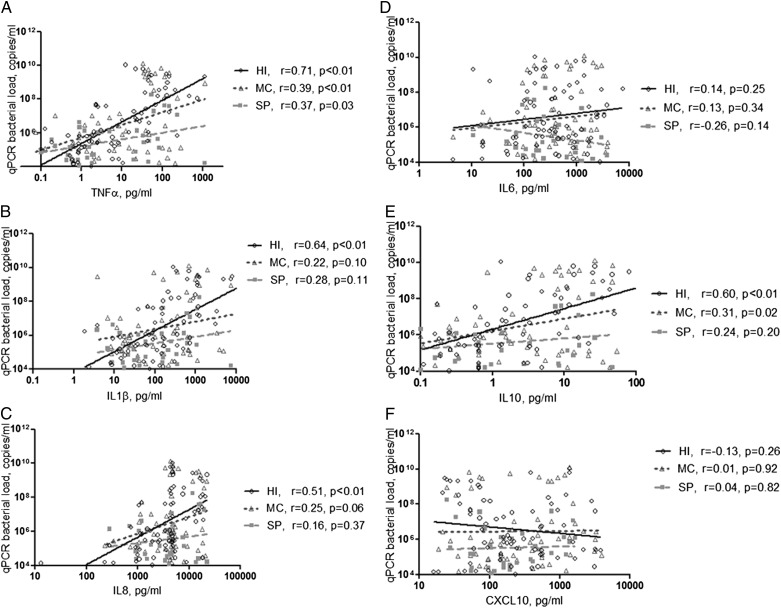
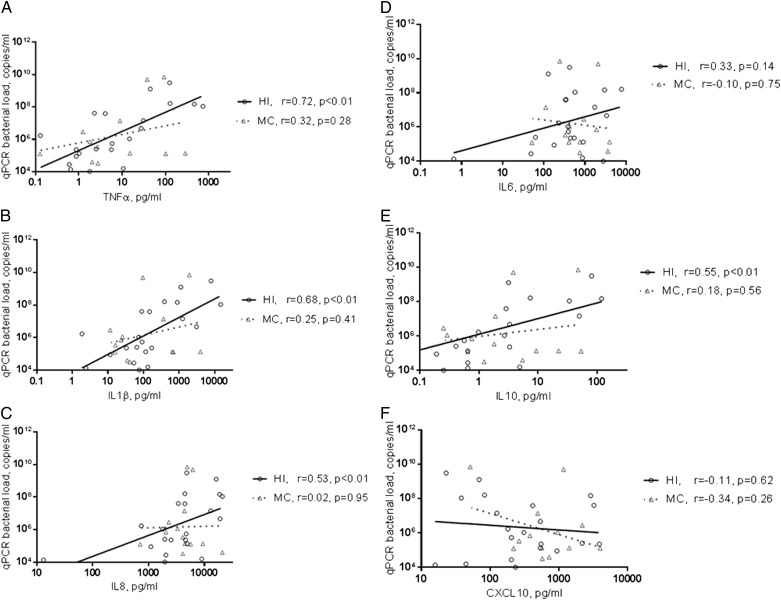
Subjects with pathogen-specific bacterial 16S qPCR PPM had higher levels of IL-1β and TNF-α and lower levels of CCL13 (Table 2, e-Table 3 (461KB, pdf) ); this was associated with a trend to a dose-response increase in inflammation with increasing numbers of qPCR PPMs (Table 3). There was no correlation of total bacterial 16S qPCR with inflammation. Pathogen-specific 16S qPCR bacterial load in subjects positive for H influenzae strongly correlated with levels of IL-1β, IL-8, IL-10, and TNF-α (r = 0.64, P < .01; r = 0.51, P < .01; r = 0.59, P < .01; and r = 0.71, P < .01, respectively). Pathogen-specific 16S qPCR bacterial load of M catarrhalis moderately correlated with IL-10 and TNF-α levels (r = 0.31, P = .02; and r = 0.39, P < .01) (Fig 2). Multivariate regression analysis identified qPCR H influenzae bacterial load and CFU as independent predictors of sputum TNF-α and IL-1β (H influenzae, β = 0.38 and 0.32 for TNF-α and IL-1β, respectively; and CFU, β = 0.36 and 0.31, respectively) (e-Table 4 (461KB, pdf) ). In subjects with either H influenzae or M catarrhalis as a single pathogen (n = 21 and n = 13, respectively), only H influenzae 16S qPCR bacterial load correlated with IL-1β, IL-10, and TNF-α (Fig 3); and only H influenzae qPCR bacterial load correlated with VAS symptoms of cough and sputum purulence and the symptom domain of the SGRQ (e-Fig 1 (461KB, pdf) ).
TABLE 2 ] .
Cytokine Levels According to Whether There Was a Pathogen-Specific Bacterial 16S qPCR PPM Detected
| Cytokine, pg/mL | qPCR PPM Negative (n = 28) | qPCR PPM Positive (n = 92) | P Value |
| IL-1β | 45 (30-68) | 142 (93-217) | < .01 |
| IL-5 | 1.3 (0.7-2.3) | 1.0 (0.7-1.4) | .58 |
| IL-6 | 292 (162-527) | 331 (236-464) | .73 |
| IL-8 | 3,104 (1,940-4,965) | 3,707 (2,916-4,712) | .49 |
| IL-10 | 0.3 (0.2-0.5) | 1.8 (1.2-2.7) | < .01 |
| TNF-α | 2.2 (1.1-4.2) | 7.2 (4.6-11.5) | < .01 |
| TNFRI | 834 (573-1,213) | 1,222 (1,003-1,489) | .07 |
| CCL2 | 605 (426-860) | 548 (441-682) | .66 |
| CCL3 | 70 (47-105) | 74 (58-94) | .83 |
| CCL4 | 957 (607-1,510) | 989 (737-1,327) | .91 |
| CCL5 | 3.2 (1.8-5.7) | 4.4 (3.5-5.5) | .23 |
| CCL13 | 47 (31-71) | 26 (20-34) | .03 |
| CCL17 | 33 (20-55) | 23 (17-30) | .21 |
| CXCL10 | 404 (260-627) | 252 (188-337) | .12 |
Data presented as geometric mean (95% CI). TNF = tumor necrosis factor. See Table 1 legend for expansion of other abbreviations.
TABLE 3 ] .
Parameters of Sputum Inflammation According to Number of qPCR Pathogens Found at Baseline
| Parameter | 0 qPCR Positive (n = 28) | 1 qPCR Positive (n = 38) | ≥ 2 qPCR Positive (n = 54) | P Value |
| IL-1β, pg/mL | 45 (30-68) | 111 (55-222) | 169 (100-287) | .01 |
| IL-5, pg/mL | 1.3 (0.7-2.3) | 1.4 (0.9-2.0) | 0.8 (0.5-1.4) | .34 |
| IL-6, pg/mL | 292 (162-527) | 473 (278-806) | 258 (168-395) | .20 |
| IL-8, pg/mL | 3,104 (1,940-4,965) | 3,578 (2,357-5,434) | 3,799 (2,851-5,064) | .77 |
| IL-10, pg/mL | 0.3 (0.2-0.5) | 1.9 (0.9-3.6) | 1.8 (1.0-3.0) | < .01 |
| TNF-α, pg/mL | 2.2 (1.1-4.2) | 5.4 (2.6-1.4) | 8.8 (4.9-15.9) | .02 |
| TNFRI, pg/mL | 843 (573-1,213) | 1,211 (903-1,623) | 1,229 (940-1,608) | .20 |
| CCL2, pg/mL | 605 (426-860) | 697 (482-1,007) | 463 (358-600) | .16 |
| CCL3, pg/mL | 70 (47-105) | 85 (56-128) | 67 (50-90) | .64 |
| CCL4, pg/mL | 957 (607-1,509) | 1,212 (793-1,851) | 858 (574-1,281) | .50 |
| CCL5, pg/mL | 3.2 (1.8-5.7) | 4.7 (3.3-6.9) | 4.2 (3.1-5.5) | .43 |
| CCL13, pg/mL | 47 (31-71) | 37 (27-51) | 21 (14-30) | < .01 |
| CCL17, pg/mL | 33 (20-55) | 31 (22-44) | 18 (12-27) | .07 |
| CXCL10, pg/mL | 404 (260-627) | 334 (211-529) | 207 (142-300) | .07 |
| Log CFU/mL, mean (95% CI) | 6.6 (5.4-7.8) | 7.3 (5.8-9.1) | 7.5 (6.0-9.1) | < .01 |
| Log 16S genome copies/mL, mean (95% CI) | 8.2 (6.8-9.4) | 8.2 (6.1-9.7) | 8.5 (6.8-10.0) | .11 |
| Total cell count, × 106/g | 2.5 (1.9-3.7) | 3.0 (1.9-4.6) | 4.1 (2.8-5.9) | .31 |
| Sputum neutrophils, mean (SEM), % | 64 (4) | 68 (4) | 75 (3) | .06 |
| Sputum neutrophil count, × 106/g | 1.6 (0.9-2.8) | 1.9 (1.1-3.2) | 2.9 (1.9-4.6) | .20 |
| Sputum eosinophils, % | 1.6 (1.1-2.2) | 1.6 (1.0-2.7) | 1.1 (0.8-1.7) | .41 |
Figure 2 –
A-F, Correlations between the sputum mediator concentrations (A) TNFα, (B) IL1β, (C) IL8, (D) IL6, (E) IL10, and (F) CXCL10 and sputum bacterial load of HI, MC, and SP. TNF = tumor necrosis factor. See Figure 1 legend for expansion of other abbreviations.
Figure 3 –
A-F, Correlations between the sputum mediator concentrations (A) TNFα, (B) IL1β, (C) IL8, (D) IL6, (E) IL10, and (F) CXCL10 and sputum bacterial load of HI and MC in the subgroup of subjects qPCR positive for a single pathogen only. See Figure 1 and 2 legends for expansion of abbreviations.
qPCR and Inflammation During an Exacerbation
Paired stable and exacerbation sputum was available in 55 subjects (men, n = 43; mean FEV1 % predicted, 51%) with an average time of sampling between stable and exacerbation of 49 days. All subjects were treated with oral antibiotic and corticosteroid therapy at the onset of an exacerbation. During an exacerbation, pathogen-specific 16S qPCR for H influenzae, M catarrhalis, and S pneumoniae were detected in 32 (58%), 26 (47%), and 20 (36%) subjects, respectively; the majority of these subjects had the same PPM qPCR at stable state (66% H influenzae, 69% M catarrhalis, and 55% S pneumoniae). In subjects who were pathogen-specific bacterial 16S qPCR PPM positive for H influenzae and M catarrhalis at exacerbation, the change in cytokine concentration between stable state and exacerbation was most marked in those who were also positive rather than negative in stable state (e-Table 5 (461KB, pdf) ). The change in pathogen-specific 16S H influenzae qPCR bacterial load between stable and exacerbation visits did not correlate with change in cytokines or change in symptoms. The change in pathogen-specific 16S M catarrhalis qPCR bacterial load between stable and exacerbation was positively correlated with a change in IL-1β and TNF-α (r = 0.37, P < .01; r = 0.31, P = .02, respectively) (e-Fig 2 (461KB, pdf) ) but was not related to changes in symptoms or health status.
Discussion
In this study we have shown that the majority of subjects with COPD at stable state had pathogenic bacteria detected by qPCR. The detection of bacteria using qPCR was associated with increased sputum IL-1β, IL-10, and TNF-α and decreased CCL13. In stable state, the strongest relationship between bacterial load, inflammation, and symptoms was observed with pathogen-specific 16S H influenzae qPCR, whether in codetection with other bacteria or as a lone pathogen. Furthermore, we determined that H influenzae qPCR bacterial load was the only pathogen that was an independent predictor of sputum TNF-α and IL-1β levels, both inflammatory chemokines. In our study, we determined that both total 16S and CFU were elevated in subjects with detectable pathogen-specific 16S but only CFU bacterial load, which suggests live bacteria growth, was independently also associated with increased inflammation. At exacerbation, we have also shown that change of M catarrhalis, but not H influenzae, bacterial load at exacerbation compared with stable state correlated with change in sputum TNF-α and IL-1β concentrations. Together, these findings suggest there is a complex dynamic relationship in COPD between bacterial load of specific pathogens, airway inflammation, and clinical expression of disease.
Our work represents the largest study to date using qPCR techniques to describe codetection in COPD.3,27,30 Prevalence of codetection was not described by Garcha et al,27 and, similar to our findings, Curran et al30 described, in a small study of 30 subjects with COPD, the presence of codetection using qPCR in 80% of subjects. Consistent with studies using culture-based15,16 and culture-independent27 techniques, we report here that H influenzae was the most commonly identified organism by qPCR. The presence of any detectable PPM was associated with increased airway inflammation and increased proinflammatory cytokines. Our data suggest that sputum TNF-α and IL-1β are more closely related to bacterial load, particularly with H influenzae, which was found to be an independent predictor of sputum TNF-α and IL-1β levels and associated with increased symptoms. Although this association does not confirm causality, there are several biologically plausible reasons that H influenzae may be of most significance. First, up-regulation of the MRLP3 inflammasome occurs during nontypeable H influenzae-induced inflammation leading to secretion of IL-1β31; second, the outer membrane protein P6 of nontypeable H influenzae has been found to induce the stimulation of TNF-α and IL-10 from human alveolar macrophages.32,33 Furthermore, alveolar macrophages have the greatest reduction of complement-independent phagocytosis of nontypeable H influenzae.34 However, it must be noted that qPCR techniques measure both live and dead bacteria. Our observation that CFU bacterial load and H influenzae 16S pathogen-specific bacterial load were closely associated with sputum levels of TNF-α and IL-1β may suggest that live H influenzae is driving this inflammation, but further studies to investigate this are warranted.
We also observed that CCL13, which is increased in eosinophilic airway disease,35 was reduced in subjects with pathogen-specific 16S qPCR PPM detection, irrespective of pathogen. Although we could not show that there is increased neutrophilic inflammation, with elevated IL8 or sputum neutrophils percent as demonstrated in previous studies,11,36 there was a trend to increase in total cell counts and absolute sputum neutrophil counts as codetection of pathogen increased and a positive correlation of IL-8 with bacterial load, further suggesting a differential airway pattern of inflammation with qPCR PPM detection. IL-10, an antiinflammatory cytokine, was also found to be increased in subjects with pathogen-specific 16S qPCR PPM and correlated with H influenzae bacterial load. Specifically, the outer membrane protein P6 of H influenzae has been shown to be a potent macrophage inducer of inflammation,33 and chronic upregulation and stimulation of macrophages may contribute to further to the pathogenesis of COPD infections.37
In contrast to earlier reports, we were unable to detect significant differences in exacerbation frequency, smoking, or inhaled corticosteroid dose between colonized and noncolonized subjects.27 However, we have demonstrated a positive association between symptoms and H influenzae bacterial load. We report here that the relationship between bacterial load and symptoms is pathogen specific and again supports a central role for sputum H influenzae bacterial load in chronic persistent disease. Whether therapy specifically targeted at reducing H influenzae bacterial load during stable state to reduce inflammatory activity in COPD is clinically beneficial is currently unknown and warrants further study.
Previous studies have demonstrated that airway inflammation increases during exacerbation episodes3,12 and that the presence of PPM during exacerbations is associated with increased IL-8, IL-1β, and TNF-α.3,38,39 However, limited data examining relationships between specific pathogens and inflammation during exacerbations exist. To our knowledge, this is the first study to explore relationships between dynamic changes in pathogen-specific bacterial load and inflammation at exacerbation. We found that change in inflammatory mediators at exacerbation compared with stable state was greatest in subjects who were pathogen-specific bacterial 16S qPCR PPM positive for either H influenzae or M catarrhalis at stable state. This suggests that there is activation of inflammation in stable state and colonized subjects with COPD, which is then further exaggerated at exacerbation. At stable state, H influenzae is closely related to airway inflammation and clinical outcomes; however, at exacerbation, changes in inflammation are more closely related to changes (including increases and decreases) of pathogen-specific bacterial 16S qPCR M catarrhalis bacterial loads. The role of this pathogen during exacerbations of COPD needs to be investigated further.
One potential limitation of this study was the use of spontaneous sputum to determine sputum inflammatory mediators and pathogen detection. Sputum collection predominately arises from the larger airways; thus, our results cannot infer causality of detectable pathogens and increased inflammation in the pathogenicity of COPD, a predominately small airways disease. However, only samples with a squamous cell contamination < 5% were used, and inflammatory mediators measured from induced and spontaneous samples have not been shown to be different,24 suggesting that the strong associations found between airway bacterial load and sputum inflammatory mediator concentrations were not clinically significant. During this study, we also selected cutoffs to determine the presence or absence of pathogen-specific 16S qPCR. Although this is an arbitrary cutoff, we acknowledge that this is in part because of a paucity of the literature, which may be missing or overdetecting pathogens, but we have used cutoffs derived from previous studies using qPCR platforms in patients with COPD.27 We have defined the presence of pathogen in sputum samples as detection and not colonization in this study, as the stable measurements were performed only at one time point, and we cannot comment on whether there is transient detection of pathogens using qPCR methods or persistence; it is clear that more work is needed to interrogate the stability and repeatability of pathogen-specific 16S qPCR in patients with COPD. A further limitation is that the presence of P aeruginosa was not assessed using qPCR techniques in this study. Although previous culture studies have suggested airway inflammation is higher in subjects in whom P aeruginosa is detected,14 its prevalence in other COPD cohorts has been low.27 Moreover, within our cohort, only five of 120 patients had grown P aeruginosa on sputum cultures within the preceding 12 months, and hence its exclusion from our analysis is unlikely to have significantly influenced our results. In our study we chose primers targeting specific pathogens in the respiratory tract, as they have previously been deemed clinically significant. However, these techniques cannot reflect the total microbiologic diversity within the airway, and newer techniques that can characterize the entire microbiome need to be studied further.40‐42 Finally, measuring sputum mediators can be fraught with difficulty, with variability in detection43‐45; however, we have only analyzed mediators that were known to be detectable using a sputum-processing method that we have previously validated in patients with airway disease.25
Conclusions
In summary, using qPCR we found that in patients with COPD, sputum pathogens are frequently detected. H influenzae was associated with increased airway inflammation and symptoms in a dose-response relationship with sputum TNF-α and IL-1β. M catarrhalis was more closely related to dynamic changes observed at exacerbation. The mechanisms by which H influenzae and M catarrhalis are related to airway inflammation in COPD warrant further investigation.
Supplementary Material
Online Supplement
Acknowledgments
Author contributions: M. B. and C. E. B. had full access to the data and are responsible for the integrity of the data and final decision to submit. B. L. B., K. H., and H. P. contributed to data collection and data interpretation; I. D. P. and M. R. B. contributed to study design and data collection and interpretation; and C. E. B. and M. B. contributed to the design of the study, volunteer recruitment, data collection, data interpretation, and data analysis; and all authors contributed to the writing of the manuscript and have approved the final version for submission.
Financial/nonfinancial disclosures: The authors have reported to CHEST the following conflicts of interest: Dr Bafadhel has received travel grants from Almirall, S.A.; Boehringer Ingelheim GmbH; and GlaxoSmithKline. Drs Pavord and Brightling have received grant support and consultancy fees from AstraZeneca, Medimmune LLC, Novartis Corporation, Roche Diagnostics, and GlaxoSmithKline. Mss Patel and Haldar and Drs Barker and Barer have reported that no potential conflicts of interest exist with any companies/organizations whose products or services may be discussed in this article.
Role of sponsors: This article presents independent research funded by the National Institute for Health Research (NIHR) and the Wellcome Trust. The views expressed are those of the authors and not necessarily those of the National Health Service, the NIHR, or the Department of Health. The Wellcome Trust, NIHR, and the European Regional Development Fund had no involvement in the design of the study, data collection, analysis, data interpretation, writing of the manuscript or the decision to submit the manuscript.
Other contributions: We thank all the volunteers who took part in the study and the following people for their assistance in subject characterization: P. Dodson, PhD; M. McCormick, BSc; S. McKenna, RGN; V. Mistry, BSc; M. Pancholi, BSc; P. Rugman, PhD; S. Terry, BSc; B. Hargadon, RGN; and A. Singapuri, BSc.
Additional information: The e-Figures and e-Tables can be found in the Supplemental Materials section of the online article.
ABBREVIATIONS
- CFU
colony-forming units
- PPM
potentially pathogenic microorganism
- qPCR
quantitative polymerase chain reaction
- SGRQ
St. George’s Respiratory Questionnaire
- TNF
tumor necrosis factor
- VAS
visual analog scale
Footnotes
Dr Barker and Ms Haldar are joint first authors and Drs Brightling and Bafadhel are joint senior authors.
FUNDING/SUPPORT: Funding was obtained from the Medical Research Council (UK), a Wellcome Senior Fellowship (C. E. B.), and a National Institute for Health Research Post-Doctoral Fellowship (M. B.), and the research was performed in laboratories partly funded by the European Regional Development Fund [Grant ERDF 05567].
References
- 1.Vestbo J, Hurd SS, Agustí AG, et al. Global strategy for the diagnosis, management, and prevention of chronic obstructive pulmonary disease: GOLD executive summary. Am J Respir Crit Care Med. 2013;187(4):347-365. [DOI] [PubMed] [Google Scholar]
- 2.Bhowmik A, Seemungal TA, Sapsford RJ, Wedzicha JA. Relation of sputum inflammatory markers to symptoms and lung function changes in COPD exacerbations. Thorax. 2000;55(2):114-120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bafadhel M, McKenna S, Terry S, et al. Acute exacerbations of chronic obstructive pulmonary disease: identification of biologic clusters and their biomarkers. Am J Respir Crit Care Med. 2011;184(6):662-671. [DOI] [PubMed] [Google Scholar]
- 4.Stockley RA, O’Brien C, Pye A, Hill SL. Relationship of sputum color to nature and outpatient management of acute exacerbations of COPD. Chest. 2000;117(6):1638-1645. [DOI] [PubMed] [Google Scholar]
- 5.Monsó E, Ruiz J, Rosell A, et al. Bacterial infection in chronic obstructive pulmonary disease. A study of stable and exacerbated outpatients using the protected specimen brush. Am J Respir Crit Care Med. 1995;152(4 pt 1):1316-1320. [DOI] [PubMed] [Google Scholar]
- 6.Banerjee D, Khair OA, Honeybourne D. Impact of sputum bacteria on airway inflammation and health status in clinical stable COPD. Eur Respir J. 2004;23(5):685-691. [DOI] [PubMed] [Google Scholar]
- 7.Sethi S, Murphy TF. Infection in the pathogenesis and course of chronic obstructive pulmonary disease. N Engl J Med. 2008;359(22):2355-2365. [DOI] [PubMed] [Google Scholar]
- 8.Papi A, Bellettato CM, Braccioni F, et al. Infections and airway inflammation in chronic obstructive pulmonary disease severe exacerbations. Am J Respir Crit Care Med. 2006;173(10):1114-1121. [DOI] [PubMed] [Google Scholar]
- 9.Zhang M, Li Q, Zhang XY, Ding X, Zhu D, Zhou X. Relevance of lower airway bacterial colonization, airway inflammation, and pulmonary function in the stable stage of chronic obstructive pulmonary disease. Eur J Clin Microbiol Infect Dis. 2010;29(12):1487-1493. [DOI] [PubMed] [Google Scholar]
- 10.Patel IS, Seemungal TA, Wilks M, Lloyd-Owen SJ, Donaldson GC, Wedzicha JA. Relationship between bacterial colonisation and the frequency, character, and severity of COPD exacerbations. Thorax. 2002;57(9):759-764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sethi S, Maloney J, Grove L, Wrona C, Berenson CS. Airway inflammation and bronchial bacterial colonization in chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2006;173(9):991-998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Aaron SD, Angel JB, Lunau M, et al. Granulocyte inflammatory markers and airway infection during acute exacerbation of chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2001;163(2):349-355. [DOI] [PubMed] [Google Scholar]
- 13.Aaron SD, Vandemheen KL, Ramsay T, et al. Multi analyte profiling and variability of inflammatory markers in blood and induced sputum in patients with stable COPD. Respir Res. 2010;11:41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hill AT, Campbell EJ, Hill SL, Bayley DL, Stockley RA. Association between airway bacterial load and markers of airway inflammation in patients with stable chronic bronchitis. Am J Med. 2000;109(4):288-295. [DOI] [PubMed] [Google Scholar]
- 15.Monsó E, Rosell A, Bonet G, et al. Risk factors for lower airway bacterial colonization in chronic bronchitis. Eur Respir J. 1999;13(2):338-342. [DOI] [PubMed] [Google Scholar]
- 16.Cabello H, Torres A, Celis R, et al. Bacterial colonization of distal airways in healthy subjects and chronic lung disease: a bronchoscopic study. Eur Respir J. 1997;10(5):1137-1144. [DOI] [PubMed] [Google Scholar]
- 17.Marin A, Garcia-Aymerich J, Sauleda J, et al. Effect of bronchial colonisation on airway and systemic inflammation in stable COPD. COPD. 2012;9(2):121-130. [DOI] [PubMed] [Google Scholar]
- 18.Anthonisen NR, Manfreda J, Warren CP, Hershfield ES, Harding GK, Nelson NA. Antibiotic therapy in exacerbations of chronic obstructive pulmonary disease. Ann Intern Med. 1987;106(2):196-204. [DOI] [PubMed] [Google Scholar]
- 19.Rodriguez-Roisin R. Toward a consensus definition for COPD exacerbations. Chest. 2000;117(5_suppl_2):398S-401S. [DOI] [PubMed] [Google Scholar]
- 20.National Clinical Guideline Centre. Chronic obstructive pulmonary disease: management of chronic obstructive pulmonary disease in adults in primary and secondary care. London, England: National Clinical Guideline Centre; 2010. [Google Scholar]
- 21.Jones PW, Quirk FH, Baveystock CM. The St George’s Respiratory Questionnaire. Respir Med. 1991;85(suppl B):25-31. [DOI] [PubMed] [Google Scholar]
- 22.Guyatt G. Measuring health status in chronic airflow limitation. Eur Respir J. 1988;1(6):560-564. [PubMed] [Google Scholar]
- 23.Brightling CE, Monterio W, Green RH, et al. Induced sputum and other outcome measures in chronic obstructive pulmonary disease: safety and repeatability. Respir Med. 2001;95(12):999-1002. [DOI] [PubMed] [Google Scholar]
- 24.Bhowmik A, Seemungal TA, Sapsford RJ, Devalia JL, Wedzicha JA. Comparison of spontaneous and induced sputum for investigation of airway inflammation in chronic obstructive pulmonary disease. Thorax. 1998;53(11):953-956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bafadhel M, McCormick M, Saha S, et al. Profiling of sputum inflammatory mediators in asthma and chronic obstructive pulmonary disease. Respiration. 2012;83(1):36-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pye A, Stockley RA, Hill SL. Simple method for quantifying viable bacterial numbers in sputum. J Clin Pathol. 1995;48(8):719-724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Garcha DS, Thurston SJ, Patel AR, et al. Changes in prevalence and load of airway bacteria using quantitative PCR in stable and exacerbated COPD. Thorax. 2012;67(12):1075-1080. [DOI] [PubMed] [Google Scholar]
- 28.Stockley RA, Bayley D, Hill SL, Hill AT, Crooks S, Campbell EJ. Assessment of airway neutrophils by sputum colour: correlation with airways inflammation. Thorax. 2001;56(5):366-372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pavord ID, Sterk PJ, Hargreave FE, et al. Clinical applications of assessment of airway inflammation using induced sputum. Eur Respir J Suppl. 2002;37:40s-43s. [DOI] [PubMed] [Google Scholar]
- 30.Curran T, Coyle PV, McManus TE, Kidney J, Coulter WA. Evaluation of real-time PCR for the detection and quantification of bacteria in chronic obstructive pulmonary disease. FEMS Immunol Med Microbiol. 2007;50(1):112-118. [DOI] [PubMed] [Google Scholar]
- 31.Rotta Detto Loria J, Rohmann K, Droemann D, et al. Nontypeable Haemophilus influenzae infection upregulates the NLRP3 inflammasome and leads to caspase-1-dependent secretion of interleukin-1β - a possible pathway of exacerbations in COPD. PLoS ONE. 2013;8(6):e66818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Berenson CS, Wrona CT, Grove LJ, et al. Impaired alveolar macrophage response to Haemophilus antigens in chronic obstructive lung disease. Am J Respir Crit Care Med. 2006;174(1):31-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Berenson CS, Murphy TF, Wrona CT, Sethi S. Outer membrane protein P6 of nontypeable Haemophilus influenzae is a potent and selective inducer of human macrophage proinflammatory cytokines. Infect Immun. 2005;73(5):2728-2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Berenson CS, Kruzel RL, Eberhardt E, Sethi S. Phagocytic dysfunction of human alveolar macrophages and severity of chronic obstructive pulmonary disease. J Infect Dis. 2013;208(12):2036-2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mendez-Enriquez E, García-Zepeda EA. The multiple faces of CCL13 in immunity and inflammation. Inflammopharmacology. 2013;21(6):397-406. [DOI] [PubMed] [Google Scholar]
- 36.Siva R, Bafadhel M, Monteiro W, Brightling CE, Pavord ID. Effect of levofloxacin on neutrophilic airway inflammation in stable COPD: a randomized, double-blind, placebo-controlled trial. Int J Chron Obstruct Pulmon Dis. 2014;9:179-186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Taylor AE, Finney-Hayward TK, Quint JK, et al. Defective macrophage phagocytosis of bacteria in COPD. Eur Respir J. 2010;35(5):1039-1047. [DOI] [PubMed] [Google Scholar]
- 38.Hurst JR, Donaldson GC, Perera WR, et al. Use of plasma biomarkers at exacerbation of chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2006;174(8):867-874. [DOI] [PubMed] [Google Scholar]
- 39.Hurst JR, Perera WR, Wilkinson TM, Donaldson GC, Wedzicha JA. Systemic and upper and lower airway inflammation at exacerbation of chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2006;173(1):71-78. [DOI] [PubMed] [Google Scholar]
- 40.Erb-Downward JR, Thompson DL, Han MK, et al. Analysis of the lung microbiome in the “healthy” smoker and in COPD. PLoS ONE. 2011;6(2):e16384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Han MK, Huang YJ, Lipuma JJ, et al. Significance of the microbiome in obstructive lung disease. Thorax. 2012;67(5):456-463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sze MA, Dimitriu PA, Hayashi S, et al. The lung tissue microbiome in chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2012;185(10):1073-1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pizzichini E, Pizzichini MM, Efthimiadis A, et al. Indices of airway inflammation in induced sputum: reproducibility and validity of cell and fluid-phase measurements. Am J Respir Crit Care Med. 1996;154(2 pt 1):308-317. [DOI] [PubMed] [Google Scholar]
- 44.Pizzichini E, Pizzichini MM, Efthimiadis A, Hargreave FE, Dolovich J. Measurement of inflammatory indices in induced sputum: effects of selection of sputum to minimize salivary contamination. Eur Respir J. 1996;9(6):1174-1180. [DOI] [PubMed] [Google Scholar]
- 45.Pizzichini MM, Popov TA, Efthimiadis A, et al. Spontaneous and induced sputum to measure indices of airway inflammation in asthma. Am J Respir Crit Care Med. 1996;154(4 pt 1):866-869. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Online Supplement