Figure 4.
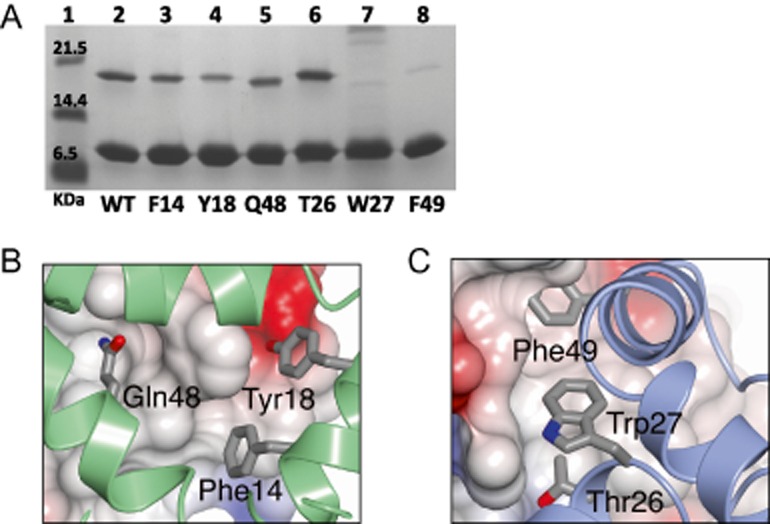
The SirA–DnaADI interface analysed by site-directed mutagenesis.
A. SDS-PAGE. Cultures of cells harbouring plasmids encoding wild type and alanine-substituted variants of His-tagged DnaADI and SirA were grown. Soluble cell lysates were prepared and loaded onto a Ni-NTA column. High imidazole eluate fractions were collected for analysis. Samples of the eluate fractions containing approximately normalized levels of DnaADI were loaded so that the efficiency of SirA pull-down could be compared. Lane 1 contains molecular weight markers. Lane 2: wild type DnaADI–SirA. Lanes 3–5: Native DnaADI and the SirA variants, loaded as follows; Lane 3: SirA(F14A), Lane 4: SirA(Y18A), Lane 5: SirA(Q48A). Lanes 6–8: Samples of native SirA and the DnaADI variants, loaded as follows; Lane 6: DnaADI(T26A), Lane 7: DnaADI(W27A), Lanes 8: DnaADI(F49A).
B and C. Core residues from the DnaA DI-interacting surface of SirA (B) and the SirA interacting surface of DnaADI (C) which were sites of alanine substitution. (B) DnaADI is shown as an electrostatic surface with SirA represented as a green ribbon with the side-chains of F14, Y18 and Q48 displayed as cylinders. (C) SirA is shown as an electrostatic surface with DnaADI represented as a blue ribbon with the side-chains of T26, W27 and F49 displayed as cylinders.
