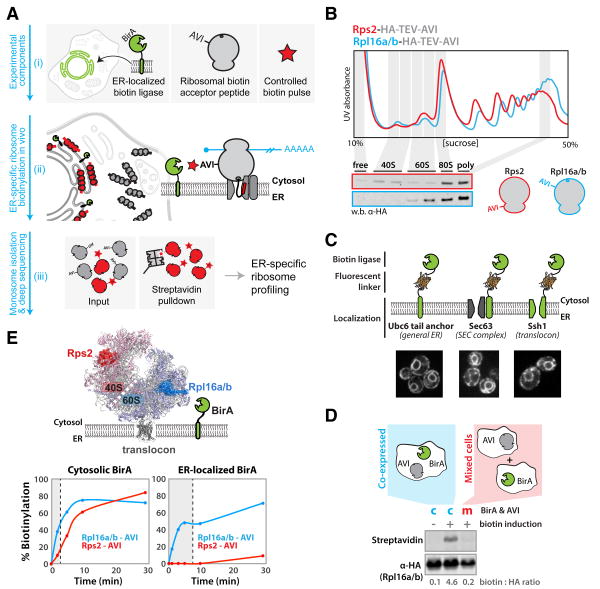
Fig. 1. A system for in vivo proximity-dependent ribosome biotinylation to monitor local protein synthesis at the ER.
(A) Schematic for proximity-specific ribosome profiling (i) The Escherichia coli biotin ligase BirA is localized to a subcellular site of interest in cells expressing an Avi-tagged ribosomal protein and grown in low-biotin conditions (ii) A biotin pulse is applied resulting in specific biotinylation of ribosomes in close physical proximity to the localized BirA (iii) Ribosome profiling of paired input (gray and red) and isolated biotinylated (red) monosomes reveals codon-resolved translational enrichment specific to the BirA locale. (B) Fractionation of yeast lysates derived from strains containing scarless C-terminal Rps2 or Rpl16a/b hemagglutinin (HA)-TEV-AviTags on 10 to 50% sucrose gradients. Polysome traces demonstrate proper ribosomal assembly and incorporation of tags into polysomes demonstrates their non-perturbative nature. (C) ER localization of BirA fusion proteins used in this study. BirA-mVenus-Ubc6, Sec63-mVenus-BirA and BirA-mVenus-Ssh1 all localize to the perinuclear and cortical ER. (D) Western blot analysis demonstrates that biotinylation of ribosomal AviTags does not occur before the addition of excess biotin or post lysis in our assay. (E) Biotinylation kinetics of 40S and 60S AviTags by BirAs localized to the cytosol or ER (Sec63). Favorable kinetics were achieved independent of localization, and preferential 60S biotinylation demonstrates the specificity of the ER-localized ligase for oriented ER ribosomes. Shaded regions indicate biotinylation times used in subsequent sequencing experiments.