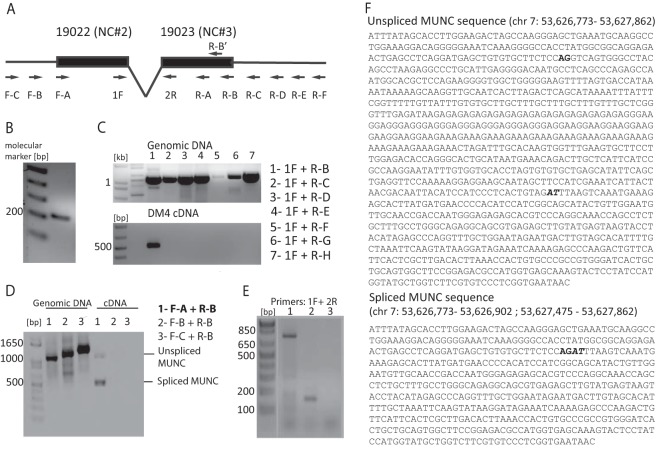
FIG 3.
Characterization of MyoD upstream noncoding transcript. (A) Schematic of primers used for 5′ RACE PCR and PCR primer walking to determine ends of MUNC transcript. (B) 5′ end mapping of MUNC: PCR product generated by 5′ RACE PCR on cap-captured DM4 C2C12 RNA with the R-B′ (nested) primer. (C) 3′ end mapping of MUNC: PCR products with 1F and indicated R primers on genomic DNA (positive control) and DM4 C2C12 cDNA. Only the R-B primer gave a product on the cDNA, putting it at the 3′ end of MUNC. (D) PCR on genomic DNA (positive control) and cDNA from DM4 C2C12 cells confirms the 5′ end of MUNC and the presence of unspliced and spliced isoforms. Primers F-A plus R-B produced two products on cDNA: genome-length unspliced (∼1,000 bases) and spliced (∼500 bases) cDNAs. (E) PCR products amplified by 1F-2R primers. Lane 1, genomic DNA from C2C12 with extension time of 60 s; lane 2, cDNA from DM3 C2C12 with an extension time of 20 s; lane 3, negative control (no DNA) with an extension time of 20 s. (F) Sequences of unspliced and spliced MUNC forms amplified by F-A and R-B primers. Bold represents the 5′ splice site, and bold italic represents the 3′ splice site. Coordinates are according to the UCSC Genome Browser (assembly of July 2007).