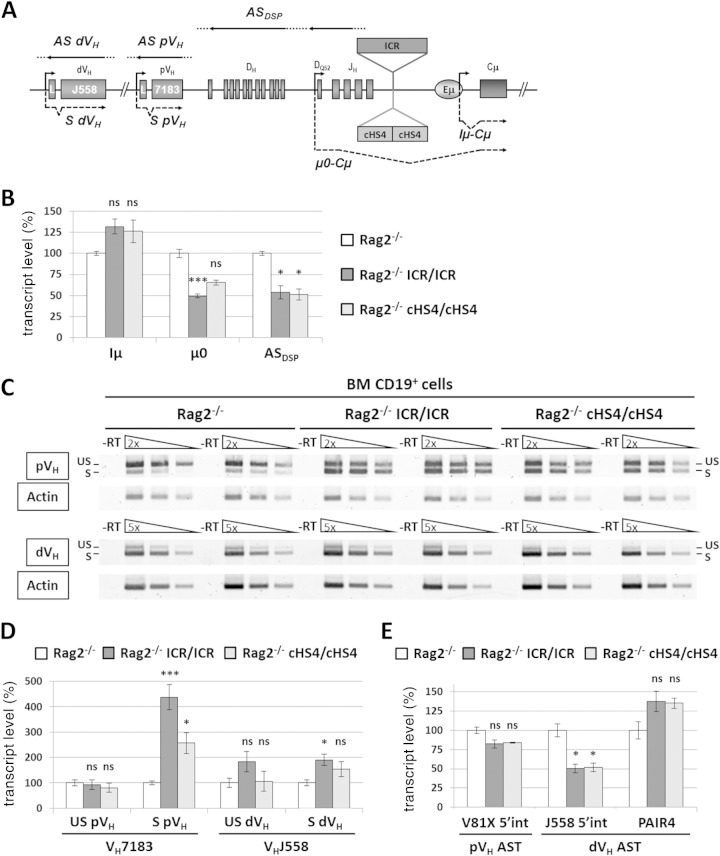
FIG 5.
Sense and antisense germ line transcription at VH genes on the mutant allele. (A) The scheme shows some of the germ line transcripts analyzed at the mutant IgH locus. Dots indicate that the initiation and termination sites of the indicated transcripts have not been mapped yet. L, leader; S, sense; AS, antisense. (B) Total RNA from sorted CD19+ cells from the bone marrow of Rag2−/−, Rag2−/− ICR/ICR, and Rag2−/− cHS4/cHS4 mice was assayed by RT-qPCR for the indicated transcripts. The corresponding transcript levels in the Rag2−/− controls were set at 100% of the signal. Gapdh (glyceraldehyde-3-phosphate dehydrogenase gene) and Ywhaz (tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein zeta gene) expression was used for normalization. The histograms show the standard errors (n ≥ 4). ***, P < 0.001; *, P < 0.05; ns, not significant. (C) Analysis of proximal VH (pVH; top) and distal VH (dVH; bottom) germ line transcripts by semiquantitative RT-PCR. Two independent samples from each genotype are shown. −RT, no reverse transcription; S, spliced transcripts; US, unspliced (antisense/primary sense) transcripts. (D) Quantification of germ line transcripts of proximal and distal VH genes by semiquantitative PCR. The signals of the corresponding transcripts in the Rag2−/− controls were set at 100%. The histograms show the standard errors (n ≥ 4). ***, P < 0.001; *, P < 0.05. (E) Quantification by qPCR of ASTs within the indicated regions of the IgH variable locus. The histograms show the standard errors (n ≥ 4). *, P < 0.05. int, intergenic.