Extended Data Fig. 7.
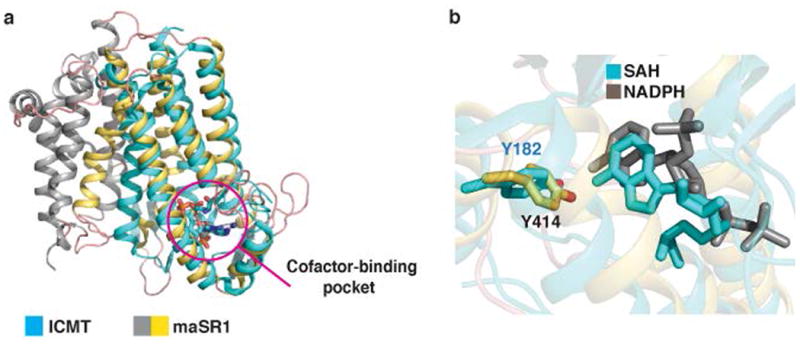
Comparison of maSR1 structure with ICMT structure. a, A comparison of maSR1 (gray and yellow) and ICMT22 (cyan) structure with SAH bound (PDB 4A2N). DALI search36 shows the closest entry (Z-score of 7.5) to maSR1 is the structure of isoprenylcysteine carboxyl methyltransferase (ICMT) consisting of 5 transmembrane helices, which had 193 Cα atoms aligned to maSR1 (TMs 6-10 and α2) with RMSD of 2.8Å. Both proteins have a similar cofactor-binding pocket (magenta circle), although the sequence conservation is low. b, Comparison of NADPH and SAH binding pockets of maSR1 (gray) and ICMT (cyan). The orientation of adenine-ribose moiety of SAH and NAPDH is similar with respect to the coordinating tyrosine residues in the cofactor pockets of these two enzymes.
